6T39
 
 | | Crystal structure of rsEGFP2 in its off-state determined by SFX | | Descriptor: | Green fluorescent protein | | Authors: | Woodhouse, J, Coquelle, N, Adam, V, Barends, T.R.M, De La Mora, E, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2019-10-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Photoswitching mechanism of a fluorescent protein revealed by time-resolved crystallography and transient absorption spectroscopy.
Nat Commun, 11, 2020
|
|
7OD3
 
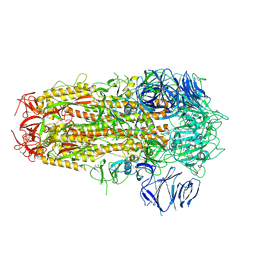 | | SARS CoV-2 Spike protein, Bristol UK Deletion variant, Closed conformation, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2021-04-28 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights in cell-type specific evolution of intra-host diversity by SARS-CoV-2.
Nat Commun, 13, 2022
|
|
1K7F
 
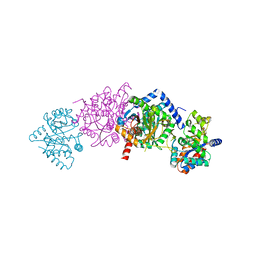 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH N-[1H-INDOL-3-YL-ACETYL]VALINE ACID | | Descriptor: | N-[1H-INDOL-3-YL-ACETYL]VALINE ACID, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN SYNTHASE ALPHA CHAIN, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-19 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a new class of allosteric effectors complexed to tryptophan synthase.
J.Biol.Chem., 277, 2002
|
|
6TC2
 
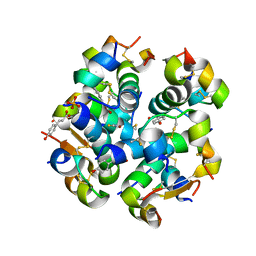 | | Monoclinic human insulin in complex with p-coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Insulin, PHOSPHATE ION, ... | | Authors: | Triandafillidis, D.-P, Parthenios, N, Spiliopoulou, M, Valmas, A, Kosinas, C, Gozzo, F, Reinle-Schmitt, M, Beckers, D, Degen, T, Pop, M, Fitch, A, Wollenhaupt, J, Weiss, M.S, Karavassili, F, Margiolaki, I. | | Deposit date: | 2019-11-04 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Insulin polymorphism induced by two polyphenols: new crystal forms and advances in macromolecular powder diffraction.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
1K8X
 
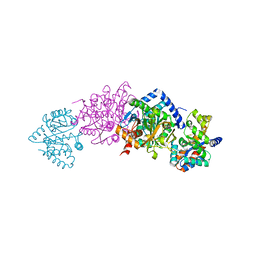 | | Crystal Structure Of AlphaT183V Mutant Of Tryptophan Synthase From Salmonella Typhimurium | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE, ... | | Authors: | Kulik, V, Weyand, M, Siedel, R, Niks, D, Arac, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2001-10-26 | | Release date: | 2002-12-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | On the Role of AlphaThr183 in the Allosteric Regulation and Catalytic Mechanism of Tryptophan Synthase
J.Mol.Biol., 324, 2002
|
|
1K3U
 
 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH N-[1H-INDOL-3-YL-ACETYL]ASPARTIC ACID | | Descriptor: | N-[1H-INDOL-3-YL-ACETYL]ASPARTIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-04 | | Release date: | 2002-07-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of a new class of allosteric effectors complexed to tryptophan synthase.
J.Biol.Chem., 277, 2002
|
|
6CMQ
 
 | | Structure of human SHP2 without N-SH2 domain | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Padua, R.A.P, Sun, Y, Marko, I, Pitsawong, W, Kern, D. | | Deposit date: | 2018-03-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of activating mutations and allosteric drug inhibition of the phosphatase SHP2.
Nat Commun, 9, 2018
|
|
1K6H
 
 | | Solution Structure of Conserved AGNN Tetraloops: Insights into Rnt1p RNA processing | | Descriptor: | RNA (5'-R(P*GP*GP*CP*GP*UP*GP*UP*UP*CP*AP*GP*AP*AP*GP*AP*AP*CP*GP*CP*GP*CP*C)-3') | | Authors: | Lebars, I, Lamontagne, B, Yoshizawa, S, Abou Elela, S, Fourmy, D. | | Deposit date: | 2001-10-16 | | Release date: | 2001-12-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of conserved AGNN tetraloops: insights into Rnt1p RNA processing.
EMBO J., 20, 2001
|
|
7NUV
 
 | |
6T46
 
 | | Structure of the Rap conjugation gene regulator of the plasmid pLS20 in complex with the Phr* peptide | | Descriptor: | CHLORIDE ION, Quorum-sensing secretion protein (processed), Response regulator aspartate phosphatase, ... | | Authors: | Crespo, I, Bernardo, N, Meijer, W.J.J, Boer, D.R. | | Deposit date: | 2019-10-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Inactivation of the dimeric RappLS20 anti-repressor of the conjugation operon is mediated by peptide-induced tetramerization.
Nucleic Acids Res., 48, 2020
|
|
6T68
 
 | | Crystal structure of Trypanosoma brucei Morn1 | | Descriptor: | CHLORIDE ION, MORN repeat-containing protein 1 | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
1K3B
 
 | | Crystal Structure of Human Dipeptidyl Peptidase I (Cathepsin C): Exclusion Domain Added to an Endopeptidase Framework Creates the Machine for Activation of Granular Serine Proteases | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, SULFATE ION, ... | | Authors: | Turk, D, Janjic, V, Stern, I, Podobnik, M, Lamba, D, Dahl, S.W, Lauritzen, C, Pedersen, J, Turk, V, Turk, B. | | Deposit date: | 2001-10-02 | | Release date: | 2002-04-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of human dipeptidyl peptidase I (cathepsin C): exclusion domain added to an endopeptidase framework creates the machine for activation of granular serine proteases.
EMBO J., 20, 2001
|
|
7NXG
 
 | | Wax synthase 1 from Acinetobacter baylyi (AbWSD1) co-crystallized with myristic acid | | Descriptor: | MYRISTIC ACID, O-acyltransferase WSD | | Authors: | Vollheyde, K, Kuehnel, K, Lambrecht, F, Kawelke, S, Herrfurth, C, Feussner, I. | | Deposit date: | 2021-03-18 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Bifunctional Wax Synthase 1 from Acinetobacter baylyi Suggests a Conformational Change upon Substrate Binding and Formation of Additional Substrate Binding Sites
Acs Catalysis, 12, 2022
|
|
1K6U
 
 | | Crystal Structure of Cyclic Bovine Pancreatic Trypsin Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Botos, I, Wu, Z, Lu, W, Wlodawer, A. | | Deposit date: | 2001-10-17 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structure of a cyclic form of bovine pancreatic trypsin inhibitor.
FEBS Lett., 509, 2001
|
|
1K7J
 
 | | Structural Genomics, protein TF1 | | Descriptor: | Protein yciO, SULFATE ION | | Authors: | Zhang, R, Dementieva, I, Thorn, J, Donnelly, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-19 | | Release date: | 2002-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Genomics, protein TF1
To be Published
|
|
6T3A
 
 | | Difference-refined structure of rsEGFP2 10 ns following 400-nm laser irradiation of the off-state determined by SFX | | Descriptor: | Green fluorescent protein | | Authors: | Woodhouse, J, Coquelle, N, Adam, V, Barends, T.R.M, De La Mora, E, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2019-10-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Photoswitching mechanism of a fluorescent protein revealed by time-resolved crystallography and transient absorption spectroscopy.
Nat Commun, 11, 2020
|
|
6T4R
 
 | | Crystal structure of Trypanosoma brucei Morn1 | | Descriptor: | MORN repeat-containing protein 1 | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
6T6O
 
 | | Y201W mutant of the orange carotenoid protein from Synechocystis at pH 4.6 | | Descriptor: | ASPARAGINE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sluchanko, N.N, Gushchin, I, Botnarevskiy, V.S, Slonimskiy, Y.B, Remeeva, A, Kovalev, K, Stepanov, A.V, Gordeliy, V, Maksimov, E.G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Role of hydrogen bond alternation and charge transfer states in photoactivation of the Orange Carotenoid Protein.
Commun Biol, 4, 2021
|
|
1K3H
 
 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
1K3L
 
 | | Crystal Structure Analysis of S-hexyl-glutathione Complex of Glutathione Transferase at 1.5 Angstroms Resolution | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1, S-HEXYLGLUTATHIONE | | Authors: | Le Trong, I, Stenkamp, R.E, Ibarra, C, Atkins, W.M, Adman, E.T. | | Deposit date: | 2001-10-03 | | Release date: | 2002-10-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.3-A resolution structure of human glutathione S-transferase with S-hexyl
glutathione bound reveals possible extended ligandin binding site
Proteins, 48, 2002
|
|
1K6D
 
 | | CRYSTAL STRUCTURE OF ACETATE COA-TRANSFERASE ALPHA SUBUNIT | | Descriptor: | ACETATE COA-TRANSFERASE ALPHA SUBUNIT, MAGNESIUM ION | | Authors: | Korolev, S, Koroleva, O, Petterson, K, Collart, F, Dementieva, I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-15 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Autotracing of Escherichia coli acetate CoA-transferase alpha-subunit structure using 3.4 A MAD and 1.9 A native data.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KA9
 
 | |
1K3G
 
 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
1K6G
 
 | | Solution Structure of Conserved AGNN Tetraloops: Insights into Rnt1p RNA Processing | | Descriptor: | RNA (5'-R(P*GP*GP*CP*GP*UP*CP*AP*UP*GP*AP*GP*UP*CP*CP*AP*UP*GP*GP*CP*GP*CP*C)-3') | | Authors: | Lebars, I, Lamontagne, B, Yoshizawa, S, Abou Elela, S, Fourmy, D. | | Deposit date: | 2001-10-16 | | Release date: | 2001-12-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of conserved AGNN tetraloops: insights into Rnt1p RNA processing.
EMBO J., 20, 2001
|
|
1K40
 
 | |
