6Z32
 
 | | Human cation-independent mannose 6-phosphate/IGF2 receptor domains 7-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-independent mannose-6-phosphate receptor, SULFATE ION, ... | | Authors: | Bochel, A.J, Williams, C, McCoy, A.J, Hoppe, H, Winter, A.J, Nicholls, R.D, Harlos, K, Jones, Y.E, Berger, I, Hassan, B, Crump, M.P. | | Deposit date: | 2020-05-19 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7-11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9.
Structure, 28, 2020
|
|
4CYM
 
 | | Complex of human VARP-ANKRD1 with Rab32-GppCp | | Descriptor: | ANKYRIN REPEAT DOMAIN-CONTAINING PROTEIN 27, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Perez-Dorado, I, Schaefer, I.B, McCoy, A.J, Owen, D.J, Evans, P.R. | | Deposit date: | 2014-04-13 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Varp is Recruited on to Endosomes by Direct Interaction with Retromer, Where Together They Function in Export to the Cell Surface.
Dev.Cell, 29, 2014
|
|
4CSJ
 
 | | The discovery of potent selective glucocorticoid receptor modulators, suitable for inhalation | | Descriptor: | 1,2-ETHANEDIOL, GLUCOCORTICOID RECEPTOR, N-[(2S)-1-[[1-(4-fluorophenyl)indazol-4-yl]amino]propan-2-yl]-2,4,6-trimethyl-benzenesulfonamide, ... | | Authors: | Edman, K, Ahlgren, R, Bengtsson, M, Bladh, H, Backstrom, S, Dahmen, J, Henriksson, K, Hillertz, P, Hulikal, V, Jerre, A, Kinchin, L, Kase, C, Lepisto, M, Mile, I, Nilsson, S, Smailagic, A, Taylor, J, Tjornebo, A, Wissler, L, Hansson, T. | | Deposit date: | 2014-03-07 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Discovery of Potent and Selective Non-Steroidal Glucocorticoid Receptor Modulators, Suitable for Inhalation.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4CVC
 
 | | Crystal structure of quinone-dependent alcohol dehydrogenase from Pseudogluconobacter saccharoketogenenes with zinc in the active site | | Descriptor: | ALCOHOL DEHYDROGENASE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rozeboom, H.J, Yu, S, Mikkelsen, R, Nikolaev, I, Mulder, H, Dijkstra, B.W. | | Deposit date: | 2014-03-25 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Quinone-Dependent Alcohol Dehydrogenase from Pseudogluconobacter Saccharoketogenes. A Versatile Dehydrogenase Oxidizing Alcohols and Carbohydrates.
Protein Sci., 24, 2015
|
|
4D0W
 
 | | Pyrrole-3-carboxamides as potent and selective JAK2 inhibitors | | Descriptor: | 5-(2-aminopyrimidin-4-yl)-2-(5-chloro-2-methylphenyl)-1H-pyrrole-3-carboxamide, GLYCEROL, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Bertrand, J, Canevari, G, Fasolini, M, Brasca, M.G, Nesi, M, Avanzi, N, Ballinari, D, Bandiera, T, Bindi, S, Carenzi, D, Casero, D, Ceriani, L, Ciomei, M, Cirla, A, Colombo, M, Cribioli, S, Cristiani, C, Della Vedova, F, Fachin, G, Felder, E.R, Galvani, A, Isacchi, A, Mirizzi, D, Motto, I, Panzeri, A, Pesenti, E, Vianello, P, Gnocchi, P, Donati, D. | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Pyrrole-3-Carboxamides as Potent and Selective Jak2 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
4D1U
 
 | | A D120A mutant of VIM-7 from Pseudomonas aeruginosa | | Descriptor: | METALLO-B-LACTAMASE, ZINC ION | | Authors: | Leiros, H.-K.S, Skagseth, S, Edvardsen, K.S.W, Lorentzen, M.S, Bjerga, G.E.K, Leiros, I, Samuelsen, O. | | Deposit date: | 2014-05-05 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | His224 Alters the R2 Drug Binding Site and Phe218 Influences the Catalytic Efficiency in the Metallo-Beta-Lactamase Vim-7.
Antimicrob.Agents Chemother., 58, 2014
|
|
6ZI3
 
 | | Crystal structure of OleP-6DEB bound to L-rhamnose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, ... | | Authors: | Montemiglio, L.C, Savino, C, Vallone, B, Parisi, G, Freda, I. | | Deposit date: | 2020-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Dissecting the Cytochrome P450 OleP Substrate Specificity: Evidence for a Preferential Substrate.
Biomolecules, 10, 2020
|
|
4CL7
 
 | | Crystal structure of VEGFR-1 domain 2 in presence of Cobalt | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 1 | | Authors: | Gaucher, J.-F, Reille-Seroussi, M, Gagey-Eilstein, N, Broussy, S, Coric, P, Seijo, B, Lascombe, M.-B, Gautier, B, Liu, W.-Q, Huguenot, F, Inguimbert, N, Bouaziz, S, Vidal, M, Broutin, I. | | Deposit date: | 2014-01-13 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biophysical Studies of the Induced Dimerization of Human Vegf R Receptor 1 Binding Domain by Divalent Metals Competing with Vegf-A
Plos One, 11, 2016
|
|
4D1S
 
 | | Pyrrole-3-carboxamides as potent and selective JAK2 inhibitors | | Descriptor: | 2-(5-chloro-2-methylphenyl)-1-methyl-5-(2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)-1H-pyrrole-3-carboxamide, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Bertrand, J, Canevari, G, Fasolini, M, Brasca, M.G, Nesi, M, Avanzi, N, Ballinari, D, Bandiera, T, Bindi, S, Carenzi, D, Casero, D, Ceriani, L, Ciomei, M, Cirla, A, Colombo, M, Cribioli, S, Cristiani, C, Della Vedova, F, Fachin, G, Felder, E.R, Galvani, A, Isacchi, A, Mirizzi, D, Motto, I, Panzeri, A, Pesenti, E, Vianello, P, Gnocchi, P, Donati, D. | | Deposit date: | 2014-05-05 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Pyrrole-3-Carboxamides as Potent and Selective Jak2 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
4D0X
 
 | | Pyrrole-3-carboxamides as potent and selective JAK2 inhibitors | | Descriptor: | 5-(2-aminopyrimidin-4-yl)-2-[2-chloro-5-(trifluoromethyl)phenyl]-1H-pyrrole-3-carboxamide, GLYCEROL, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Canevari, G, Fasolini, M, Bertrand, J, Brasca, M.G, Nesi, M, Avanzi, N, Ballinari, D, Bandiera, T, Bindi, S, Carenzi, D, Casero, D, Ceriani, L, Ciomei, M, Cirla, A, Colombo, M, Cribioli, S, Cristiani, C, Della Vedova, F, Fachin, G, Felder, E.R, Galvani, A, Isacchi, A, Mirizzi, D, Motto, I, Panzeri, A, Pesenti, E, Vianello, P, Gnocchi, P, Donati, D. | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Pyrrole-3-Carboxamides as Potent and Selective Jak2 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
4D1T
 
 | | High resolution structure of native tVIM-7 from Pseudomonas aeruginosa | | Descriptor: | METALLO-B-LACTAMASE, ZINC ION | | Authors: | Leiros, H.-K.S, Skagseth, S, Edvardsen, K.S.W, Lorentzen, M.S, Bjerga, G.E.K, Leiros, I, Samuelsen, O. | | Deposit date: | 2014-05-05 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | His224 Alters the R2 Drug Binding Site and Phe218 Influences the Catalytic Efficiency in the Metallo-Beta-Lactamase Vim-7.
Antimicrob.Agents Chemother., 58, 2014
|
|
4D60
 
 | | Structure of a dimeric Plasmodium falciparum profilin mutant | | Descriptor: | PROFILIN, SULFATE ION | | Authors: | Bhargav, S.P, Vahokoski, J, Kallio, J.P, Torda, A, Kursula, P, Kursula, I. | | Deposit date: | 2014-11-07 | | Release date: | 2015-06-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Two Independently Folding Units of Plasmodium Profilin Suggest Evolution Via Gene Fusion.
Cell.Mol.Life Sci., 72, 2015
|
|
4D6X
 
 | | Crystal structure of the receiver domain of NtrX from Brucella abortus | | Descriptor: | BACTERIAL REGULATORY, FIS FAMILY PROTEIN, IMIDAZOLE | | Authors: | Klinke, S, Fernandez, I, Carrica, M.C, Otero, L.H, Goldbaum, F.A. | | Deposit date: | 2014-11-18 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Snapshots of Conformational Changes Shed Light Into the Ntrx Receiver Domain Signal Transduction Mechanism
J.Mol.Biol., 427, 2015
|
|
4D1V
 
 | | A F218Y mutant of VIM-7 from Pseudomonas aeruginosa | | Descriptor: | METALLO-B-LACTAMASE, ZINC ION | | Authors: | Leiros, H.-K.S, Skagseth, S, Edvardsen, K.S.W, Lorentzen, M.S, Bjerga, G.E.K, Leiros, I, Samuelsen, O. | | Deposit date: | 2014-05-05 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | His224 Alters the R2 Drug Binding Site and Phe218 Influences the Catalytic Efficiency in the Metallo-Beta-Lactamase Vim-7.
Antimicrob.Agents Chemother., 58, 2014
|
|
4D2G
 
 | | Crystal structure of human PCNA in complex with p15 peptide | | Descriptor: | P15, PROLIFERATING CELL NUCLEAR ANTIGEN | | Authors: | DeBiasio, A, Ibanez, A, Mortuza, G, Molina, R, Cordeiro, T.N, Castillo, F, Villate, M, Merino, N, Lelli, M, Diercks, T, Luque, I, Bernardo, P, Montoya, G, Blanco, F.J. | | Deposit date: | 2014-05-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of P15(Paf)-PCNA Complex and Implications for Clamp Sliding During DNA Replication and Repair.
Nat.Commun., 6, 2015
|
|
4D3G
 
 | | Structure of PstA | | Descriptor: | PSTA | | Authors: | Campeotto, I, Freemont, P, Grundling, A. | | Deposit date: | 2014-10-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Complex Structure and Biochemical Characterization of the Staphylococcus Aureus Cyclic Di-AMP Binding Protein Psta, the Founding Member of a New Signal Transduction Protein Family
J.Biol.Chem., 290, 2015
|
|
4CLV
 
 | | Crystal Structure of dodecylphosphocholine-solubilized NccX from Cupriavidus metallidurans 31A | | Descriptor: | NICKEL-COBALT-CADMIUM RESISTANCE PROTEIN NCCX, PHOSPHATE ION, PHOSPHOCHOLINE, ... | | Authors: | Legrand, P, Girard, E, Petit-Hartlein, I, Maillard, A.P, Coves, J. | | Deposit date: | 2014-01-15 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | The X-Ray Structure of Nccx from Cupriavidus Metallidurans 31A Illustrates Potential Dangers of Detergent Solubilization When Generating and Interpreting Crystal Structures of Membrane Proteins.
J.Biol.Chem., 289, 2014
|
|
4CVD
 
 | |
4CXF
 
 | | Structure of CnrH in complex with the cytosolic domain of CnrY | | Descriptor: | CHLORIDE ION, CNRY, RNA POLYMERASE SIGMA FACTOR CNRH, ... | | Authors: | Maillard, A.P, Girard, E, Ziani, W, Petit-Hartlein, I, Kahn, R, Coves, J. | | Deposit date: | 2014-04-07 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Crystal Structure of the Anti-Sigma Factor Cnry in Complex with the Sigma Factor Cnrh Shows a New Structural Class of Anti- Sigma Factors Targeting Extracytoplasmic-Function Sigma Factors.
J.Mol.Biol., 426, 2014
|
|
4E1P
 
 | |
6OCP
 
 | | Crystal structure of a human GABAB receptor peptide bound to KCTD16 T1 | | Descriptor: | BTB/POZ domain-containing protein KCTD16, Gamma-aminobutyric acid type B receptor subunit 2 | | Authors: | Zuo, H, Glaaser, I, Zhao, Y, Kurinov, I, Mosyak, L, Wang, H, Liu, J, Park, J, Frangaj, A, Sturchler, E, Zhou, M, McDonald, P, Geng, Y, Slesinger, P.A, Fan, Q.R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for auxiliary subunit KCTD16 regulation of the GABABreceptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OCR
 
 | | Crystal structure of human KCTD16 T1 domain | | Descriptor: | BTB/POZ domain-containing protein KCTD16 | | Authors: | Zuo, H, Glaaser, I, Zhao, Y, Kurinov, I, Mosyak, L, Wang, H, Liu, J, Park, J, Frangaj, A, Sturchler, E, Zhou, M, McDonald, P, Geng, Y, Slesinger, P.A, Fan, Q.R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis for auxiliary subunit KCTD16 regulation of the GABABreceptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OCT
 
 | | Crystal structure of human KCTD16 T1 domain | | Descriptor: | BTB/POZ domain-containing protein KCTD16 | | Authors: | Zuo, H, Glaaser, I, Zhao, Y, Kurinov, I, Mosyak, L, Wang, H, Liu, J, Park, J, Frangaj, A, Sturchler, E, Zhou, M, McDonald, P, Geng, Y, Slesinger, P.A, Fan, Q.R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for auxiliary subunit KCTD16 regulation of the GABABreceptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6PH4
 
 | | Full length LOV-PAS-HK construct from the LOV-HK sensory protein from Brucella abortus (light-adapted, construct 15-489) | | Descriptor: | Blue-light-activated histidine kinase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Rinaldi, J, Otero, L.H, Fernandez, I, Goldbaum, F.A, Shin, H, Yang, X, Klinke, S. | | Deposit date: | 2019-06-25 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
7O3E
 
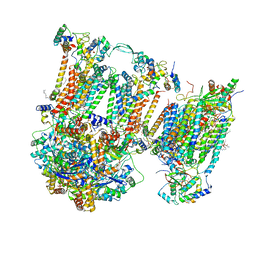 | | Murine supercomplex CIII2CIV in the intermediate locked conformation | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2021-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and assembly of the mammalian mitochondrial supercomplex CIII 2 CIV.
Nature, 598, 2021
|
|
