8EIN
 
 | |
8F4H
 
 | | RT XFEL structure of Photosystem II 1200 microseconds after the third illumination at 2.10 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4F
 
 | | RT XFEL structure of Photosystem II 500 microseconds after the third illumination at 2.03 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4C
 
 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.00 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8EZ5
 
 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.09 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-10-31 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4K
 
 | | RT XFEL structure of the three-flash state of Photosystem II (3F, S0-rich) at 2.16 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4I
 
 | | RT XFEL structure of Photosystem II 2000 microseconds after the third illumination at 2.00 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4D
 
 | | RT XFEL structure of Photosystem II 50 microseconds after the third illumination at 2.15 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4E
 
 | | RT XFEL structure of Photosystem II 250 microseconds after the third illumination at 2.09 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4G
 
 | | RT XFEL structure of Photosystem II 730 microseconds after the third illumination at 2.03 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4J
 
 | | RT XFEL structure of Photosystem II 4000 microseconds after the third illumination at 2.00 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
5YDV
 
 | | Regulatory domain of HypT from Salmonella typhimurium complexed with HOCl (HOCl-bound form) | | Descriptor: | Cell density-dependent motility repressor, SULFATE ION, hypochlorous acid | | Authors: | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | Deposit date: | 2017-09-14 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5YEZ
 
 | | Regulatory domain of HypT M206Q mutant from Salmonella typhimurium | | Descriptor: | Cell density-dependent motility repressor | | Authors: | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | Deposit date: | 2017-09-20 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5YDW
 
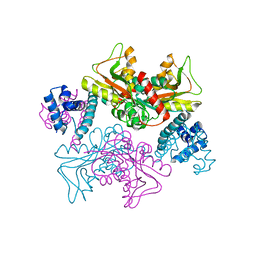 | | Full-length structure of HypT from Salmonella typhimuriuma (hypochlorite-specific LysR-type transcriptional regulator) | | Descriptor: | Cell density-dependent motility repressor | | Authors: | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | Deposit date: | 2017-09-15 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8C2H
 
 | | Transmembrane domain of active state homomeric GluA1 AMPA receptor in tandem with TARP gamma 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 1 flip isoform, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8C1P
 
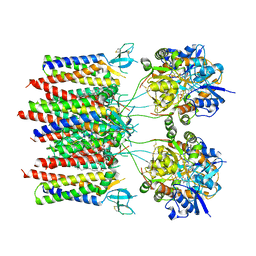 | | Active state homomeric GluA1 AMPA receptor in complex with TARP gamma 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CYCLOTHIAZIDE, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8C1S
 
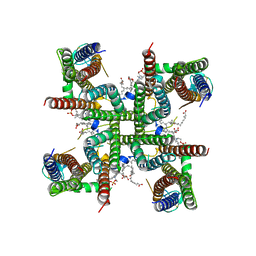 | | Transmembrane domain of resting state homomeric GluA2 F231A mutant AMPA receptor in complex with TARP gamma 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 2, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8C2I
 
 | | Transmembrane domain of resting state homomeric GluA1 AMPA receptor in complex with TARP gamma 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 1 flip isoform, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8C1R
 
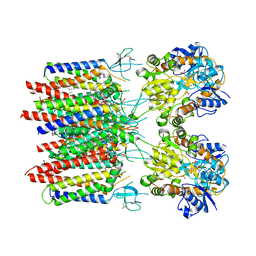 | | Resting state homomeric GluA2 F231A mutant AMPA receptor in complex with TARP gamma-2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 2, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8C1Q
 
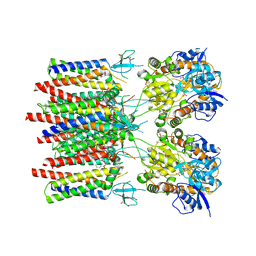 | | Resting state homomeric GluA1 AMPA receptor in complex with TARP gamma 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 1 flip isoform, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8EYM
 
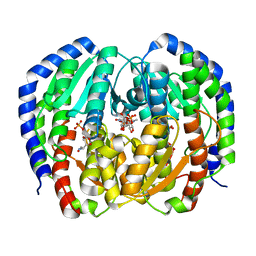 | | CRYSTAL STRUCTURE OF NAGB-II PHOSPHOSUGAR ISOMERASE FROM SHEWANELLA DENITRIFICANS OS217 IN COMPLEX WITH GLUCITOLAMINE-6-PHOSPHATE AND N-ACETYLGLUCOSAMINE-6-PHOSPHATE AT 2.31 A RESOLUTION | | Descriptor: | 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, GLUCOSAMINE-6-PHOSPHATE DEAMINASE | | Authors: | Rodriguez-Hernandez, A, Marcos-Viquez, J, Rodriguez-Romero, A, Bustos-Jaimes, I. | | Deposit date: | 2022-10-27 | | Release date: | 2023-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | Substrate binding in the allosteric site mimics homotropic cooperativity in the SIS-fold glucosamine-6-phosphate deaminases.
Protein Sci., 32, 2023
|
|
8C4Y
 
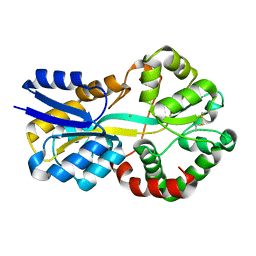 | | SFX structure of FutA bound to Fe(III) | | Descriptor: | FE (III) ION, Putative iron ABC transporter, substrate binding protein | | Authors: | Bolton, R, Tews, I. | | Deposit date: | 2023-01-05 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A redox switch allows binding of Fe(II) and Fe(III) ions in the cyanobacterial iron-binding protein FutA from Prochlorococcus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8EOL
 
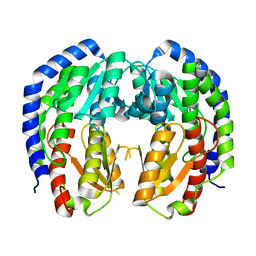 | |
5NZS
 
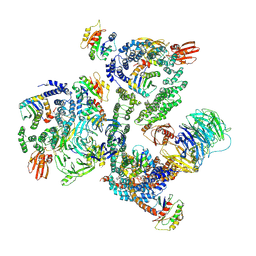 | | The structure of the COPI coat leaf in complex with the ArfGAP2 uncoating factor | | Descriptor: | ADP-ribosylation factor 1, ADP-ribosylation factor GTPase-activating protein 2, Coatomer subunit alpha, ... | | Authors: | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (10.1 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
8F2R
 
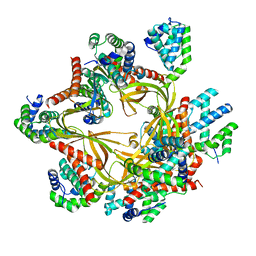 | | Human CCC complex | | Descriptor: | COMM domain-containing protein 1, COMM domain-containing protein 10, COMM domain-containing protein 2, ... | | Authors: | Healy, M.D, McNally, K.E, Butkovic, R, Chilton, M, Kato, K, Sacharz, J, McConville, C, Moody, E.R.R, Shaw, S, Planelles-Herrero, V.J, Kadapalakere, S.Y, Ross, J, Borucu, U, Palmer, C.S, Chen, K, Croll, T.I, Hall, R.J, Caruana, N.J, Ghai, R, Nguyen, T.H.D, Heesom, K.J, Saitoh, S, Berger, I, Berger-Schaffitzel, C, Williams, T.A, Stroud, D.A, Derivery, E, Collins, B.M, Cullen, P.J. | | Deposit date: | 2022-11-08 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structure of the endosomal Commander complex linked to Ritscher-Schinzel syndrome.
Cell, 186, 2023
|
|
