5OEX
 
 | | Complex with iodine ion for thiocyanate dehydrogenase from Thioalkalivibrio paradoxus | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Tsallagov, S.I, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2017-07-10 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and characterization of a novel copper containing enzyme - THIOCYANATE DEHYDROGENASE.
To Be Published
|
|
8DGG
 
 | | Structure of glycosylated LAG-3 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Silberstein, J.L, Mathews, I.I, Frank, J.A, Chan, K.-W, Fernandez, D, Du, J, Wang, J, Kong, X.-P, Cochran, J.R. | | Deposit date: | 2022-06-23 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.78 Å) | | Cite: | Structural insights reveal interplay between LAG-3 homodimerization, ligand binding, and function.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6I4I
 
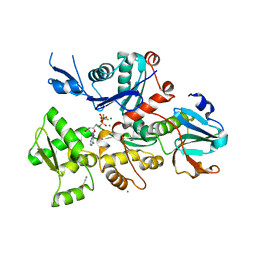 | | Crystal Structure of Plasmodium falciparum actin I (F54Y mutant) in the Mg-K-ADP-AlFn state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Actin-1, ... | | Authors: | Kumpula, E.-P, Lopez, A.J, Tajedin, L, Han, H, Kursula, I. | | Deposit date: | 2018-11-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Atomic view into Plasmodium actin polymerization, ATP hydrolysis, and fragmentation.
Plos Biol., 17, 2019
|
|
6P5N
 
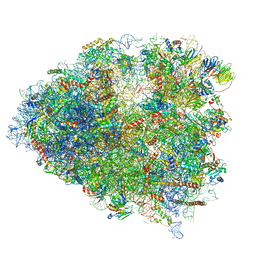 | | Structure of a mammalian 80S ribosome in complex with a single translocated Israeli Acute Paralysis Virus IRES and eRF1 | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
3JTM
 
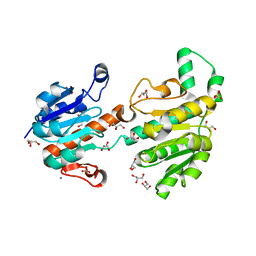 | | Structure of recombinant formate dehydrogenase from Arabidopsis thaliana | | Descriptor: | AZIDE ION, Formate dehydrogenase, mitochondrial, ... | | Authors: | Timofeev, V.I, Shabalin, I.G, Serov, A.E, Polyakov, K.M, Popov, V.O, Tishkov, V.I, Kuranova, I.P, Samigina, V.R. | | Deposit date: | 2009-09-13 | | Release date: | 2010-09-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of recombinant formate dehydrogenase from Arabidopsis thaliana
to be published
|
|
6ZMZ
 
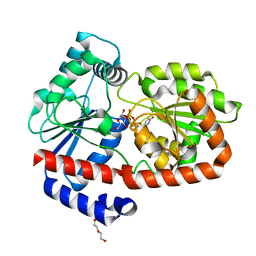 | | UDPG-bound Trehalose transferase from Thermoproteus uzoniensis | | Descriptor: | DI(HYDROXYETHYL)ETHER, Trehalose phosphorylase/synthase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Bento, I, Mestrom, L, Marsden, S.R, van der Eijk, H, Laustsen, J.U, Jeffries, C.M, Svergun, D.I, Hagedoorn, P.-H, Hanefeld, U. | | Deposit date: | 2020-07-05 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Anomeric Selectivity of Trehalose Transferase with Rare l-Sugars.
Acs Catalysis, 10, 2020
|
|
5JGP
 
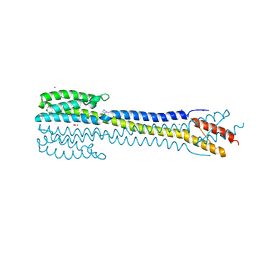 | | Crystal structure of the nitrate/nitrite sensor NarQ fragment bound with iodide ions | | Descriptor: | IODIDE ION, NITRATE ION, Nitrate/nitrite sensor protein NarQ | | Authors: | Melnikov, I, Polovinkin, V, Popov, A, Gordeliy, V. | | Deposit date: | 2016-04-20 | | Release date: | 2017-05-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fast iodide-SAD phasing for high-throughput membrane protein structure determination.
Sci Adv, 3, 2017
|
|
6OCB
 
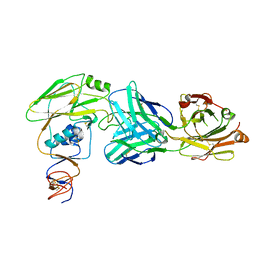 | |
6ZQJ
 
 | | Cryo-EM structure of trimeric prME spike of Spondweni virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, prM | | Authors: | Renner, M, Dejnirattisai, W, Carrique, L, Serna Martin, I, Karia, D, Ilca, S.L, Ho, S.F, Kotecha, A, Keown, J.R, Mongkolsapaya, J, Screaton, G.R, Grimes, J.M. | | Deposit date: | 2020-07-09 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Flavivirus maturation leads to the formation of an occupied lipid pocket in the surface glycoproteins.
Nat Commun, 12, 2021
|
|
1GDJ
 
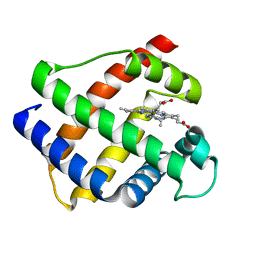 | |
8QCF
 
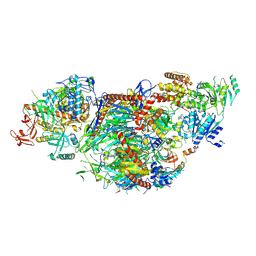 | | yeast cytoplasmic exosome-Ski2 complex degrading a RNA substrate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Antiviral helicase SKI2, Exosome complex component CSL4, ... | | Authors: | Keidel, A, Koegel, A, Reichelt, P, Kowalinski, E, Schaefer, I.B, Conti, E. | | Deposit date: | 2023-08-25 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.
Mol.Cell, 83, 2023
|
|
8QJR
 
 | | BRG1 bromodomain in complex with VBC via compound 17 | | Descriptor: | (2S,4R)-1-[(2R)-2-[3-[2-[4-[3-[4-[(1R,5S)-3-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]pyridin-2-yl]oxycyclobutyl]oxypiperidin-1-yl]ethoxy]-1,2-oxazol-5-yl]-3-methyl-butanoyl]-N-[(1S)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, CHLORIDE ION, Elongin-B, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
8QJT
 
 | | BRM (SMARCA2) Bromodomain in complex with ligand 10 | | Descriptor: | 2-[6-azanyl-5-[(1R,5S)-8-[2-(2-methoxyethoxy)pyridin-4-yl]-3,8-diazabicyclo[3.2.1]octan-3-yl]pyridazin-3-yl]phenol, CHLORIDE ION, Probable global transcription activator SNF2L2, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.568 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
7AT8
 
 | | Histone H3 recognition by nucleosome-bound PRC2 subunit EZH2. | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3.2, ... | | Authors: | Finogenova, K, Benda, C, Schaefer, I.B, Poepsel, S, Strauss, M, Mueller, J. | | Deposit date: | 2020-10-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis for PRC2 decoding of active histone methylation marks H3K36me2/3.
Elife, 9, 2020
|
|
6SOF
 
 | | human insulin receptor ectodomain bound by 4 insulin | | Descriptor: | Insulin, Insulin receptor | | Authors: | Gutmann, T, Schaefer, I.B, Poojari, C.S, Vattulainen, I, Strauss, M, Coskun, U. | | Deposit date: | 2019-08-29 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of the complete and ligand-saturated insulin receptor ectodomain.
J.Cell Biol., 219, 2020
|
|
6PLD
 
 | | Crystal Structure of Pseudomonas aeruginosa D-Arginine Dehydrogenase Y249F variant with 6-OH-FAD - Green fraction | | Descriptor: | 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, FAD-dependent catabolic D-arginine dehydrogenase DauA, ... | | Authors: | Reis, R.A.G, Iyer, A, Agniswamy, J, Gannavaram, S, Weber, I, Gadda, G. | | Deposit date: | 2019-06-30 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Single-Point Mutation in d-Arginine Dehydrogenase Unlocks a Transient Conformational State Resulting in Altered Cofactor Reactivity.
Biochemistry, 60, 2021
|
|
6I4E
 
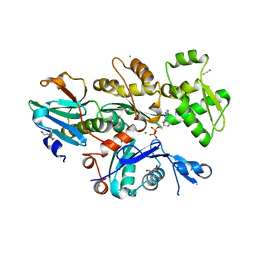 | | Crystal Structure of Plasmodium falciparum actin I in the Mg-ADP state | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Actin-1, ... | | Authors: | Kumpula, E.-P, Lopez, A.J, Tajedin, L, Han, H, Kursula, I. | | Deposit date: | 2018-11-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Atomic view into Plasmodium actin polymerization, ATP hydrolysis, and fragmentation.
Plos Biol., 17, 2019
|
|
6OBZ
 
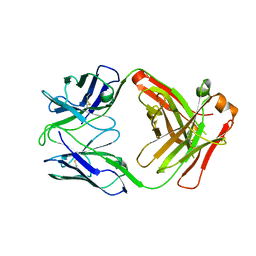 | | Crystal structure of FluA-20 Fab | | Descriptor: | Heavy chain of FluA-20 Fab, Light chain of FluA-20 Fab, Immunoglobulin light chain | | Authors: | Wilson, I.A, Lang, S. | | Deposit date: | 2019-03-21 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A Site of Vulnerability on the Influenza Virus Hemagglutinin Head Domain Trimer Interface.
Cell, 177, 2019
|
|
7VGX
 
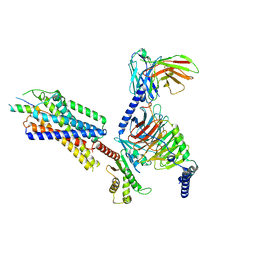 | | Neuropeptide Y Y1 Receptor (NPY1R) in Complex with G Protein and its endogeneous Peptide-Agonist Neuropeptide Y (NPY) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Park, C, Kim, J, Jeong, H, Kang, H, Bang, I, Choi, H.-J. | | Deposit date: | 2021-09-19 | | Release date: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of neuropeptide Y signaling through Y1 receptor
Nat Commun, 13, 2022
|
|
6VEJ
 
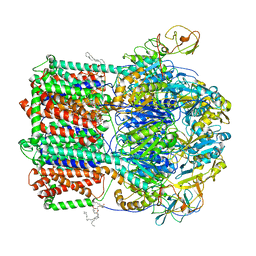 | | TriABC transporter from Pseudomonas aeruginosa | | Descriptor: | (2R)-2-(hexadecanoyloxy)propyl nonadecanoate, DODECYL-ALPHA-D-MALTOSIDE, PALMITIC ACID, ... | | Authors: | Sygusch, J, Fabre, L, Rouiller, I, Bhattacharyya, S. | | Deposit date: | 2020-01-02 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | A "Drug Sweeping" State of the TriABC Triclosan Efflux Pump from Pseudomonas aeruginosa.
Structure, 29, 2021
|
|
8QU4
 
 | | NF-YB/C Heterodimer in Complex with a 13-mer NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker in an alternative binding pose | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nuclear transcription factor Y subunit alpha, ... | | Authors: | Arbore, F, Durukan, C, Klintrot, C.I.R, Grossmann, T.N, Hennig, S. | | Deposit date: | 2023-10-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Binding Dynamics of a Stapled Peptide Targeting the Transcription Factor NF-Y.
Chembiochem, 25, 2024
|
|
8QU3
 
 | | NF-YB/C Heterodimer in Complex with a 13-mer NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, Nuclear transcription factor Y subunit alpha, ... | | Authors: | Arbore, F, Durukan, C, Klintrot, C.I.R, Grossmann, T.N, Hennig, S. | | Deposit date: | 2023-10-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Binding Dynamics of a Stapled Peptide Targeting the Transcription Factor NF-Y.
Chembiochem, 25, 2024
|
|
7QX1
 
 | | ATAD2 in complex with FragLite7 | | Descriptor: | 1,2-ETHANEDIOL, 4-iodanyl-3~{H}-pyridin-2-one, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-01-26 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
8QQ1
 
 | | SpNOX dehydrogenase domain, mutant F397W in complex with Flavin adenine dinucleotide (FAD) | | Descriptor: | BROMIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Oxidoreductase | | Authors: | Humm, A.S, Dupeux, F, Vermot, A, Petit-Harleim, I, Fieschi, F, Marquez, J.A. | | Deposit date: | 2023-10-03 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | X-ray structure and enzymatic study of a bacterial NADPH oxidase highlight the activation mechanism of eukaryotic NOX.
Elife, 13, 2024
|
|
4YNM
 
 | | ASH1L wild-type SET domain in complex with S-adenosyl methionine (SAM) | | Descriptor: | Histone-lysine N-methyltransferase ASH1L, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Rogawski, D.S, Ndoj, J, Cho, H.-J, Maillard, I, Grembecka, J, Cierpicki, T. | | Deposit date: | 2015-03-10 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Two Loops Undergoing Concerted Dynamics Regulate the Activity of the ASH1L Histone Methyltransferase.
Biochemistry, 54, 2015
|
|
