3X2P
 
 | | Neutron and X-ray joint refined structure of PcCel45A with cellopentaose at 298K. | | Descriptor: | Endoglucanase V-like protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Tanaka, I, Niimura, N, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-14 | | Last modified: | 2020-07-29 | | Method: | NEUTRON DIFFRACTION (1.518 Å), X-RAY DIFFRACTION | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X0Q
 
 | | ADP ribose pyrophosphatase from Thermus thermophilus HB8 in ESMM-state at reaction time of 20 min | | Descriptor: | ADP-ribose pyrophosphatase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Furuike, Y, Akita, Y, Miyahara, I, Kamiya, N. | | Deposit date: | 2014-10-17 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | ADP-Ribose Pyrophosphatase Reaction in Crystalline State Conducted by Consecutive Binding of Two Manganese(II) Ions as Cofactors
Biochemistry, 55, 2016
|
|
3X2O
 
 | | Neutron and X-ray joint refined structure of PcCel45A apo form at 298K. | | Descriptor: | Endoglucanase V-like protein | | Authors: | Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Tanaka, I, Niimura, N, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-07 | | Last modified: | 2019-12-18 | | Method: | NEUTRON DIFFRACTION (1.5 Å), X-RAY DIFFRACTION | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3RXD
 
 | | Crystal structure of Trypsin complexed with (3-methoxyphenyl)methanamine | | Descriptor: | 1-(3-methoxyphenyl)methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3X0I
 
 | | ADP ribose pyrophosphatase in apo state at 0.91 angstrom resolution | | Descriptor: | ADP-ribose pyrophosphatase, GLYCEROL, SULFATE ION | | Authors: | Furuike, Y, Akita, Y, Miyahara, I, Kamiya, N. | | Deposit date: | 2014-10-16 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | ADP-Ribose Pyrophosphatase Reaction in Crystalline State Conducted by Consecutive Binding of Two Manganese(II) Ions as Cofactors
Biochemistry, 55, 2016
|
|
3RXB
 
 | | Crystal structure of Trypsin complexed with 4-guanidinobutanoic acid | | Descriptor: | 4-carbamimidamidobutanoic acid, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3X02
 
 | | Crystal structure of PIP4KIIBETA complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
2NZ2
 
 | | Crystal structure of human argininosuccinate synthase in complex with aspartate and citrulline | | Descriptor: | ASPARTIC ACID, Argininosuccinate synthase, CITRULLINE, ... | | Authors: | Karlberg, T, Uppenberg, J, Arrowsmith, C, Berglund, H, Busam, R.D, Collins, R, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Hogbom, M, Johansson, I, Kotenyova, T, Magnusdottir, A, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Wallden, K, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-22 | | Release date: | 2006-12-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human argininosuccinate synthetase.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3X0N
 
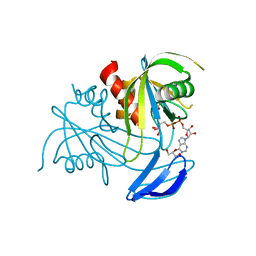 | | ADP ribose pyrophosphatase from Thermus thermophilus HB8 in ESM-state at reaction time of 6 min | | Descriptor: | ADP-ribose pyrophosphatase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Furuike, Y, Akita, Y, Miyahara, I, Kamiya, N. | | Deposit date: | 2014-10-17 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | ADP-Ribose Pyrophosphatase Reaction in Crystalline State Conducted by Consecutive Binding of Two Manganese(II) Ions as Cofactors
Biochemistry, 55, 2016
|
|
4HH2
 
 | | Structure of PpsR without the HTH motif from Rb. sphaeroides | | Descriptor: | Transcriptional regulator, PpsR | | Authors: | Winkler, A, Heintz, U, Lindner, R, Reinstein, J, Shoeman, R, Schlichting, I. | | Deposit date: | 2012-10-09 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A ternary AppA-PpsR-DNA complex mediates light regulation of photosynthesis-related gene expression.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4AFD
 
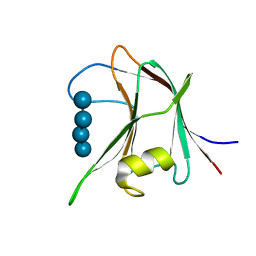 | | Structural and biochemical characterization of a novel Carbohydrate Binding Module of endoglucanase Cel5A from Eubacterium cellulosolvens with a partially bound cellotetraose moeity. | | Descriptor: | ENDOGLUCANASE CEL5A, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-01-18 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Understanding How Noncatalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan.
J.Biol.Chem., 288, 2013
|
|
4HEG
 
 | | Crystal Structure of HIV-1 protease mutants R8Q complexed with inhibitor GRL-0519 | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, HIV-1 protease | | Authors: | Zhang, H, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2012-10-03 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1 protease.
J.Med.Chem., 56, 2013
|
|
4GQO
 
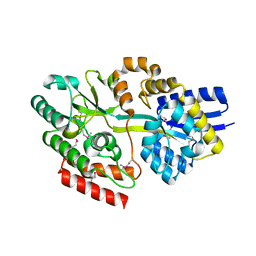 | | 2.1 Angstrom resolution crystal structure of uncharacterized protein lmo0859 from Listeria monocytogenes EGD-e | | Descriptor: | Lmo0859 protein, TRIETHYLENE GLYCOL | | Authors: | Halavaty, A.S, Filippova, E.V, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-23 | | Release date: | 2012-09-19 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom resolution crystal structure of uncharacterized protein lmo0859 from Listeria monocytogenes EGD-e
To be Published
|
|
4A92
 
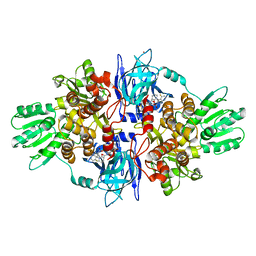 | | Full-length HCV NS3-4A protease-helicase in complex with a macrocyclic protease inhibitor. | | Descriptor: | (1'R,2R,2'S,6S,24AS)-17-FLUORO-6-(1-METHYL-2-OXOPIPERIDINE-3-CARBOXAMIDO)-19,19-DIOXIDO-5,21,24-TRIOXO-2'-VINYL-1,2,3,5,6,7,8,9,10,11,12,13,14,20,21,23,24,24A-OCTADECAHYDROSPIRO[BENZO[S]PYRROLO[2,1-G][1,2,5,8,18]THIATETRAAZACYCLOICOSINE-22,1'-CYCLOPRO-2-CARBOXYLATEPAN]-2-YL 4-FLUOROISOINDOLINE, SERINE PROTEASE NS3, ZINC ION | | Authors: | Schiering, N, D'Arcy, A, Simic, O, Eder, J, Raman, P, Svergun, D.I, Bodendorf, U. | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | A Macrocyclic Hcv Ns3/4A Protease Inhibitor Interacts with Protease and Helicase Residues in the Complex with its Full- Length Target.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4GYE
 
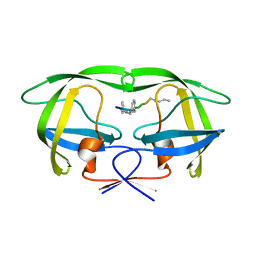 | | MDR 769 HIV-1 Protease in Complex with Reduced P1F | | Descriptor: | P1F peptide, Protease | | Authors: | Dewdney, T.G, Wang, Y, Brunzelle, J, Reiter, S.J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2012-09-05 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Ligand modifications to reduce the relative resistance of multi-drug resistant HIV-1 protease.
Bioorg.Med.Chem., 21, 2013
|
|
3TNJ
 
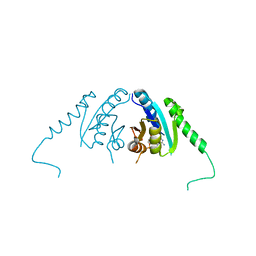 | | Crystal structure of universal stress protein from Nitrosomonas europaea with AMP bound | | Descriptor: | ADENOSINE MONOPHOSPHATE, Universal stress protein (Usp) | | Authors: | Tkaczuk, K.L, Chruszcz, M, Shumilin, I.A, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-01 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional insight into the universal stress protein family.
Evol Appl, 6, 2013
|
|
4H4E
 
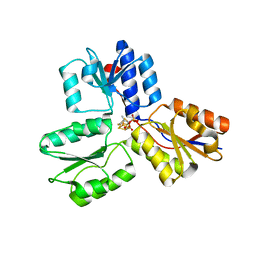 | | IspH in complex with (E)-4-mercapto-3-methylbut-2-enyl diphosphate | | Descriptor: | (2E)-3-methyl-4-sulfanylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Span, I, Eisenreich, W, Jauch, J, Bacher, A, Groll, M. | | Deposit date: | 2012-09-17 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Fluoro, Amino, and Thiol Inhibitors Bound to the [Fe(4) S(4) ] Protein IspH.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
3TPK
 
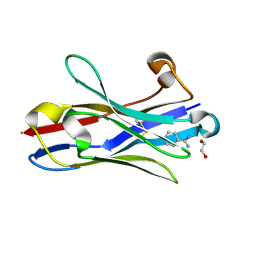 | | Crystal structure of the oligomer-specific KW1 antibody fragment | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDINE, Immunoglobulin heavy chain antibody variable domain KW1 | | Authors: | Parthier, C, Morgado, I, Stubbs, M.T, Fandrich, M. | | Deposit date: | 2011-09-08 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis of beta-amyloid oligomer recognition with a conformational antibody fragment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UR3
 
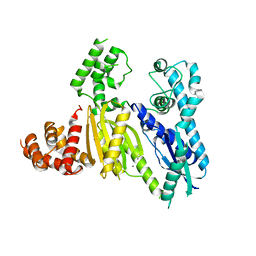 | | Structure of the Cmr2 subunit of the CRISPR RNA silencing complex | | Descriptor: | CALCIUM ION, Cmr2dHD, ZINC ION | | Authors: | Cocozaki, A.I, Ramia, N.F, Shao, Y, Hale, C.R, Terns, R.M, Terns, M.P, Li, H. | | Deposit date: | 2011-11-21 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Structure of the Cmr2 Subunit of the CRISPR-Cas RNA Silencing Complex.
Structure, 20, 2012
|
|
4H7Z
 
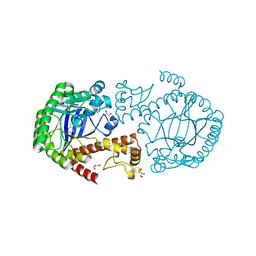 | | tRNA-guanine transglycosylase Y106F, C158V mutant in complex with guanine | | Descriptor: | GLYCEROL, GUANINE, Queuine tRNA-ribosyltransferase, ... | | Authors: | Biela, I, Tidten-Luksch, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor.
Plos One, 8, 2013
|
|
3UT2
 
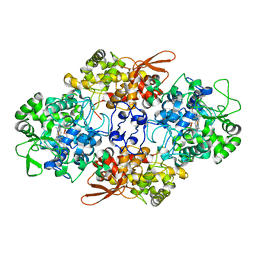 | | Crystal Structure of Fungal MagKatG2 | | Descriptor: | Catalase-peroxidase 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zamocky, M, Garcia-Fernandez, M.Q, Gasselhuber, B, Jakopitsch, C, Furtmuller, P.G, Loewen, P.C, Fita, I, Obinger, C, Carpena, X. | | Deposit date: | 2011-11-24 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | High Conformational Stability of Secreted Eukaryotic Catalase-peroxidases: ANSWERS FROM FIRST CRYSTAL STRUCTURE AND UNFOLDING STUDIES.
J.Biol.Chem., 287, 2012
|
|
3V4G
 
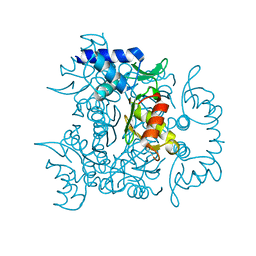 | | 1.60 Angstrom resolution crystal structure of an arginine repressor from Vibrio vulnificus CMCP6 | | Descriptor: | Arginine repressor | | Authors: | Halavaty, A.S, Minasov, G, Filippova, E, Shuvalova, L, Winsor, J, Dubrovska, I, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.60 Angstrom resolution crystal structure of an arginine repressor from Vibrio vulnificus CMCP6
To be Published
|
|
3U2I
 
 | | X-ray crystal structure of human Transthyretin at room temperature | | Descriptor: | Transthyretin | | Authors: | Yokoyama, T, Mizuguchi, M, Nabeshima, Y, Kusaka, K, Yamada, T, Hosoya, T, Ohhara, T, Kurihara, K, Tomoyori, K, Tanaka, I, Niimura, N. | | Deposit date: | 2011-10-03 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrogen-bond network and pH sensitivity in transthyretin: Neutron crystal structure of human transthyretin
J.Struct.Biol., 177, 2012
|
|
4HDB
 
 | | Crystal Structure of HIV-1 protease mutants D30N complexed with inhibitor GRL-0519 | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, HIV-1 Protease, ... | | Authors: | Zhang, H, Wang, Y.-F, Shen, C.H, Agniswamy, J, Weber, I.T. | | Deposit date: | 2012-10-02 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1 protease.
J.Med.Chem., 56, 2013
|
|
3UVD
 
 | | Crystal Structure of the bromodomain of human Transcription activator BRG1 (SMARCA4) in complex with N-Methyl-2-pyrrolidone | | Descriptor: | 1-methylpyrrolidin-2-one, Transcription activator BRG1 | | Authors: | Filippakopoulos, P, Felletar, I, Picaud, S, Keates, T, Muniz, J, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-29 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
