4GSK
 
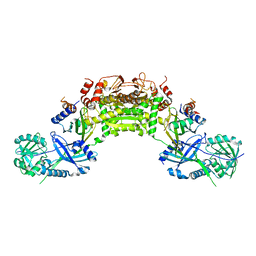 | | Crystal structure of an Atg7-Atg10 crosslinked complex | | Descriptor: | Ubiquitin-like modifier-activating enzyme ATG7, Ubiquitin-like-conjugating enzyme ATG10, ZINC ION | | Authors: | Kaiser, S.E, Mao, K, Taherbhoy, A.M, Yu, S, Olszewski, J.L, Duda, D.M, Kurinov, I, Deng, A, Fenn, T.D, Klionsky, D.J, Schulman, B.A. | | Deposit date: | 2012-08-27 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Noncanonical E2 recruitment by the autophagy E1 revealed by Atg7-Atg3 and Atg7-Atg10 structures.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3T7G
 
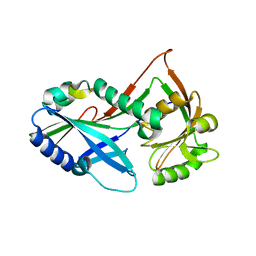 | | Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway | | Descriptor: | Autophagy-related protein 3, Ubiquitin-like modifier-activating enzyme ATG7 | | Authors: | Taherbhoy, A.M, Tait, S.W, Kaiser, S.E, Williams, A.H, Deng, A, Nourse, A, Hammel, M, Kurinov, I, Rock, C.O, Green, D.R, Schulman, B.A. | | Deposit date: | 2011-07-30 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Atg8 transfer from atg7 to atg3: a distinctive e1-e2 architecture and mechanism in the autophagy pathway.
Mol.Cell, 44, 2011
|
|
3WRE
 
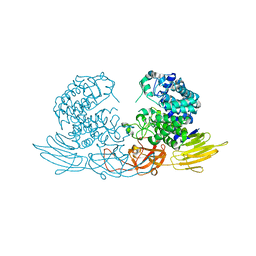 | | The crystal structure of native HypBA1 from Bifidobacterium longum JCM 1217 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
3WW0
 
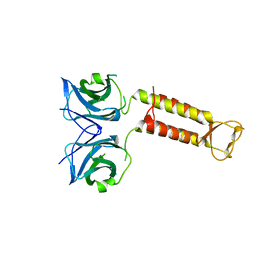 | | Crystal structure of F97A mutant, a new nuclear transport receptor of Hsp70 | | Descriptor: | Protein Hikeshi | | Authors: | Song, J, Kose, S, Watanabe, A, Son, S.Y, Choi, S, Hong, R.H, Yamashita, E, Park, I.Y, Imamoto, N, Lee, S.J. | | Deposit date: | 2014-06-12 | | Release date: | 2015-03-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional analysis of Hikeshi, a new nuclear transport receptor of Hsp70s
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WEJ
 
 | | Crystal structure of the human squalene synthase F288A mutant in complex with presqualene pyrophosphate | | Descriptor: | MAGNESIUM ION, Squalene synthase, {(1R,2R,3R)-2-[(3E)-4,8-dimethylnona-3,7-dien-1-yl]-2-methyl-3-[(1E,5E)-2,6,10-trimethylundeca-1,5,9-trien-1-yl]cyclopropyl}methyl trihydrogen diphosphate | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H.J. | | Deposit date: | 2013-07-07 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the catalytic mechanism of human squalene synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WBJ
 
 | | Crystal structure analysis of eukaryotic translation initiation factor 5B structure II | | Descriptor: | Eukaryotic translation initiation factor 5B | | Authors: | Zheng, A, Yamamoto, R, Ose, T, Yu, J, Tanaka, I, Yao, M. | | Deposit date: | 2013-05-20 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | X-ray structures of eIF5B and the eIF5B-eIF1A complex: the conformational flexibility of eIF5B is restricted on the ribosome by interaction with eIF1A
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3TBH
 
 | |
3WIK
 
 | | Crystal structure of the CK2alpha/compound10 complex | | Descriptor: | Casein kinase II subunit alpha, N-[5-(4-nitrophenyl)-1,3,4-thiadiazol-2-yl]acetamide | | Authors: | Kinoshita, T, Nakaniwa, T, Sekiguchi, Y, Nakanishi, I. | | Deposit date: | 2013-09-18 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Identification of protein kinase CK2 inhibitors using solvent dipole ordering virtual screening
To be Published
|
|
3WRG
 
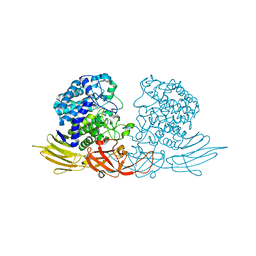 | | The complex structure of HypBA1 with L-arabinose | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase, ZINC ION, beta-L-arabinofuranose | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
4GZO
 
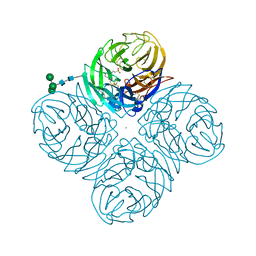 | | N2 neuraminidase of A/Tanzania/205/2010 H3N2 in complex with hepes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2012-09-06 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Influenza virus neuraminidases with reduced enzymatic activity that avidly bind sialic Acid receptors.
J.Virol., 86, 2012
|
|
4H4N
 
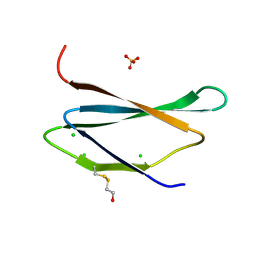 | | 1.1 Angstrom Crystal Structure of Hypothetical Protein BA_2335 from Bacillus anthracis | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-17 | | Release date: | 2012-09-26 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | 1.1 Angstrom Crystal Structure of Hypothetical Protein BA_2335 from Bacillus anthracis.
TO BE PUBLISHED
|
|
3WEG
 
 | | Crystal structure of the human squalene synthase in complex with farnesyl thiopyrophosphate and magnesium ion | | Descriptor: | MAGNESIUM ION, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE, Squalene synthase | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H.J. | | Deposit date: | 2013-07-07 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the catalytic mechanism of human squalene synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WRF
 
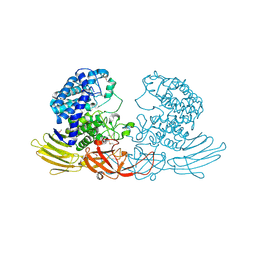 | | The crystal structure of native HypBA1 from Bifidobacterium longum JCM 1217 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
3RJ5
 
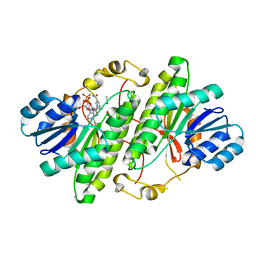 | | Structure of alcohol dehydrogenase from Drosophila lebanonesis T114V mutant complexed with NAD+ | | Descriptor: | ACETIC ACID, Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Morgunova, E, Wuxiuer, Y, Cols, N, Popov, A, Sylte, I, Karshikoff, A, Gonzales-Duarte, R, Ladenstein, R, Winberg, J.O. | | Deposit date: | 2011-04-15 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | An intact eight-membered water chain in drosophilid alcohol dehydrogenases is essential for optimal enzyme activity.
Febs J., 279, 2012
|
|
3ZJ0
 
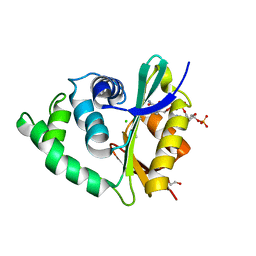 | | The human O-GlcNAcase C-terminal domain is a pseudo histone acetyltransferase | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, ACETYLTRANSFERASE, ... | | Authors: | Rao, F.V, Schuettelkopf, A.W, Dorfmueller, H.C, Ferenbach, A.T, Navratilova, I, van Aalten, D.M.F. | | Deposit date: | 2013-01-15 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Bacterial Putative Acetyltransferase Defines the Fold of the Human O-Glcnacase C-Terminal Domain.
Open Biol., 3, 2013
|
|
4IH2
 
 | | Crystal structure of kirola (Act d 11) from crystal soaked with 2-aminopurine | | Descriptor: | CHLORIDE ION, Kirola, UNKNOWN LIGAND | | Authors: | Chruszcz, M, Ciardiello, M.A, Giangrieco, I, Osinski, T, Minor, W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and bioinformatic analysis of the kiwifruit allergen Act d 11, a member of the family of ripening-related proteins.
Mol.Immunol., 56, 2013
|
|
3RFA
 
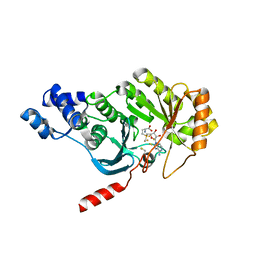 | | X-ray structure of RlmN from Escherichia coli in complex with S-adenosylmethionine | | Descriptor: | IRON/SULFUR CLUSTER, Ribosomal RNA large subunit methyltransferase N, S-ADENOSYLMETHIONINE | | Authors: | Boal, A.K, Grove, T.L, McLaughlin, M.I, Yennawar, N, Booker, S.J, Rosenzweig, A.C. | | Deposit date: | 2011-04-05 | | Release date: | 2011-05-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for methyl transfer by a radical SAM enzyme.
Science, 332, 2011
|
|
4IHP
 
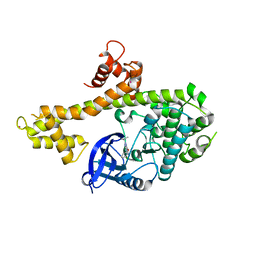 | | Crystal structure of TgCDPK1 with inhibitor bound | | Descriptor: | 1-tert-butyl-3-(3-chlorophenoxy)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-domain protein kinase 1, UNKNOWN ATOM OR ION | | Authors: | El Bakkouri, M, Tempel, W, Crandall, I.E, Massad, T, Loppnau, P, Graslund, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kain, C.K, Shokat, K.M, Sibley, L.D, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-19 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of TgCDPK1 with inhibitor bound
TO BE PUBLISHED
|
|
5UAB
 
 | |
3RQ9
 
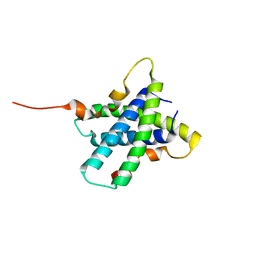 | | Structure of Tsi2, a Tse2-immunity protein from Pseudomonas aeruginosa | | Descriptor: | Type VI secretion immunity protein | | Authors: | Li, M, Le Trong, I, Stenkamp, R.E, Mougous, J.D. | | Deposit date: | 2011-04-27 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural Basis for Type VI Secretion Effector Recognition by a Cognate Immunity Protein.
Plos Pathog., 8, 2012
|
|
4IFO
 
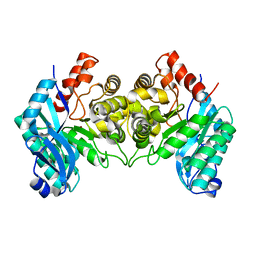 | | 2.50 Angstroms X-ray crystal structure of R51A 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase from Pseudomonas fluorescens | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, ZINC ION | | Authors: | Huo, L, Davis, I, Chen, L, Liu, A. | | Deposit date: | 2012-12-14 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The power of two: arginine 51 and arginine 239* from a neighboring subunit are essential for catalysis in alpha-amino-beta-carboxymuconate-epsilon-semialdehyde decarboxylase.
J.Biol.Chem., 288, 2013
|
|
4IG2
 
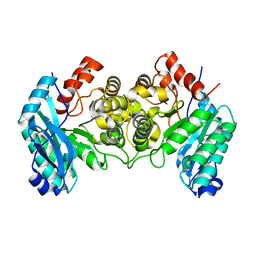 | | 1.80 Angstroms X-ray crystal structure of R51A and R239A heterodimer 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase from Pseudomonas fluorescens | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, ZINC ION | | Authors: | Huo, L, Davis, I, Chen, L, Liu, A. | | Deposit date: | 2012-12-15 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The power of two: arginine 51 and arginine 239* from a neighboring subunit are essential for catalysis in alpha-amino-beta-carboxymuconate-epsilon-semialdehyde decarboxylase.
J.Biol.Chem., 288, 2013
|
|
3RXT
 
 | | Crystal structure of Trypsin complexed with (3-methoxyphenyl)methanamin (F04 and F03, cocktail experiment) | | Descriptor: | 1-(3-methoxyphenyl)methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3ZN8
 
 | | Structural Basis of Signal Sequence Surveillance and Selection by the SRP-SR Complex | | Descriptor: | 4.5 S RNA, DIPEPTIDYL AMINOPEPTIDASE B, MAGNESIUM ION, ... | | Authors: | von Loeffelholz, O, Knoops, K, Ariosa, A, Zhang, X, Karuppasamy, M, Huard, K, Schoehn, G, Berger, I, Shan, S.O, Schaffitzel, C. | | Deposit date: | 2013-02-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural Basis of Signal Sequence Surveillance and Selection by the Srp-Sr Complex
Nat.Struct.Mol.Biol., 20, 2013
|
|
3S0D
 
 | | Apis mellifera OBP 14 in complex with the citrus odorant citralva (3,7-dimethylocta-2,6-dienenitrile) | | Descriptor: | (2Z)-3,7-dimethylocta-2,6-dienenitrile, OBP14 | | Authors: | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
