1GPJ
 
 | | Glutamyl-tRNA Reductase from Methanopyrus kandleri | | Descriptor: | (2R,3R,4S,5S)-4-AMINO-2-[6-(DIMETHYLAMINO)-9H-PURIN-9-YL]-5-(HYDROXYMETHYL)TETRAHYDRO-3-FURANOL, CITRIC ACID, GLUTAMIC ACID, ... | | Authors: | Moser, J, Schubert, W.-D, Beier, V, Bringemeier, I, Jahn, D, Heinz, D.W. | | Deposit date: | 2001-11-05 | | Release date: | 2002-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | V-shaped structure of glutamyl-tRNA reductase, the first enzyme of tRNA-dependent tetrapyrrole biosynthesis.
EMBO J., 20, 2001
|
|
3O55
 
 | | Crystal structure of human FAD-linked augmenter of liver regeneration (ALR) | | Descriptor: | Augmenter of liver regeneration, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Banci, L, Bertini, I, Calderone, V, Cefaro, C, Ciofi-Baffoni, S, Gallo, A, Kallergi, E, Lionaki, E, Pozidis, C, Tokatlidis, K. | | Deposit date: | 2010-07-28 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular recognition and substrate mimicry drive the electron-transfer process between MIA40 and ALR.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4MKR
 
 | | Structure of the apo form of a Zingiber officinale double bond reductase | | Descriptor: | Zingiber officinale double bond reductase | | Authors: | Buratto, J, Langlois d'Estaintot, B, Granier, T, Gallois, B, Willis, M.A, Sang, Y, Flores-Sanchez, I.J, Gang, D.R. | | Deposit date: | 2013-09-05 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of Zingiber officinale double bond reductase
To be Published
|
|
1ZUU
 
 | | Crystal structure of the yeast Bzz1 first SH3 domain at 0.97-A resolution | | Descriptor: | BZZ1 protein, MAGNESIUM ION, UNKNOWN ATOM OR ION | | Authors: | Kursula, P, Kursula, I, Lehmann, F, Zou, P, Song, Y.H, Wilmanns, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Structural genomics of yeast SH3 domains
To be Published
|
|
4Q7F
 
 | | 1.98 Angstrom Crystal Structure of Putative 5'-Nucleotidase from Staphylococcus aureus in complex with Adenosine. | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, 5' nucleotidase family protein, MAGNESIUM ION, ... | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-04-24 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | 1.98 Angstrom Crystal Structure of Putative 5'-Nucleotidase from Staphylococcus aureus in complex with Adenosine.
TO BE PUBLISHED
|
|
7LY4
 
 | | Cryo-EM structure of the elongation module of the bacillamide NRPS, BmdB, in complex with the oxidase, BmdC | | Descriptor: | BmdB, bacillamide NRPS, BmdC, ... | | Authors: | Sharon, I, Fortinez, C.M, Schmeing, T.M. | | Deposit date: | 2021-03-05 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures and function of a tailoring oxidase in complex with a nonribosomal peptide synthetase module.
Nat Commun, 13, 2022
|
|
3NQ3
 
 | | Bovine beta-lactoglobulin complex with capric acid | | Descriptor: | Beta-lactoglobulin, CHLORIDE ION, DECANOIC ACID, ... | | Authors: | Loch, J.I, Lewinski, K. | | Deposit date: | 2010-06-29 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Two modes of fatty acid binding to bovine beta-lactoglobulin-crystallographic and spectroscopic studies
J.Mol.Recognit., 24, 2011
|
|
1OHB
 
 | | Acetylglutamate kinase from Escherichia coli complexed with ADP and sulphate | | Descriptor: | ACETATE ION, ACETYLGLUTAMATE KINASE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gil-Ortiz, F, Ramon-Maiques, S, Fita, I, Rubio, V. | | Deposit date: | 2003-05-23 | | Release date: | 2003-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Course of Phosphorus in the Reaction of N-Acetyl-L-Glutamate Kinase, Determined from the Structures of Crystalline Complexes, Including a Complex with an Alf(4)(-) Transition State Mimic
J.Mol.Biol., 331, 2003
|
|
7LY7
 
 | | Crystal structure of the elongation module of the bacillamide NRPS, BmdB, in complex with the oxidase BmdC | | Descriptor: | 5'-{[(2R,3S)-3-amino-2-({2-[(N-{(2R)-4-[(dihydroxyphosphanyl)oxy]-2-hydroxy-3,3-dimethylbutanoyl}-beta-alanyl)amino]ethyl}sulfanyl)-4-sulfanylbutane-1-sulfonyl]amino}-5'-deoxyadenosine, BmdB, Bacillamide NRPS, ... | | Authors: | Fortinez, C.M, Sharon, I, Schmeing, T.M. | | Deposit date: | 2021-03-05 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structures and function of a tailoring oxidase in complex with a nonribosomal peptide synthetase module.
Nat Commun, 13, 2022
|
|
3OAI
 
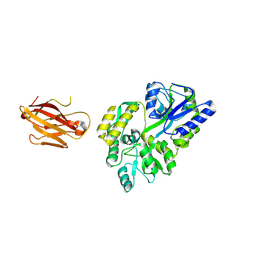 | | Crystal structure of the extra-cellular domain of human myelin protein zero | | Descriptor: | Maltose-binding periplasmic protein, Myelin protein P0, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Sohi, J, Kamholz, J, Kovari, L.C. | | Deposit date: | 2010-08-05 | | Release date: | 2011-12-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the extracellular domain of human myelin protein zero.
Proteins, 80, 2012
|
|
3NTL
 
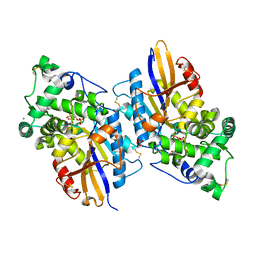 | |
1ZW7
 
 | | Elimination of the C-cap in Ubiquitin Structure, Dynamics and Thermodynamic Consequences | | Descriptor: | Ubiquitin | | Authors: | Ermolenko, D.N, Dangi, B, Gronenborn, A.M, Makhatadze, G.I. | | Deposit date: | 2005-06-03 | | Release date: | 2006-05-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Elimination of the C-cap in ubiquitin-structure, dynamics and thermodynamic consequences.
Biophys.Chem., 126, 2007
|
|
2XF4
 
 | | Crystal structure of Salmonella enterica serovar typhimurium YcbL | | Descriptor: | HYDROXYACYLGLUTATHIONE HYDROLASE, TETRAETHYLENE GLYCOL, ZINC ION | | Authors: | Stamp, A, Owen, P, El Omari, K, Nichols, C, Lockyer, M, Lamb, H, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2010-05-20 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Characterization of Salmonella Enterica Serovar Typhimurium Ycbl: An Unusual Type II Glyoxalase
Protein Sci., 19, 2010
|
|
1OH9
 
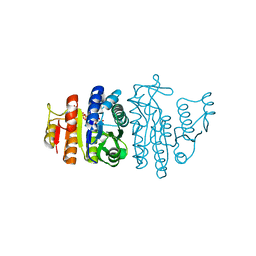 | | Acetylglutamate kinase from Escherichia coli complexed with MgADP, N-acetyl-L-glutamate and the transition-state mimic AlF4- | | Descriptor: | ACETYLGLUTAMATE KINASE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gil-Ortiz, F, Ramon-Maiques, S, Fita, I, Rubio, V. | | Deposit date: | 2003-05-23 | | Release date: | 2003-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The Course of Phosphorus in the Reaction of N-Acetyl-L-Glutamate Kinase, Determined from the Structures of Crystalline Complexes, Including a Complex with an Alf(4)(-) Transition State Mimic
J.Mol.Biol., 331, 2003
|
|
3O1A
 
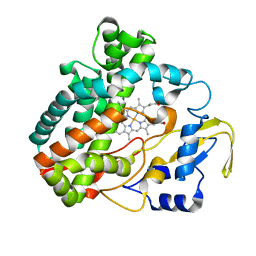 | |
210L
 
 | | PROTEIN STRUCTURE PLASTICITY EXEMPLIFIED BY INSERTION AND DELETION MUTANTS IN T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, T4 LYSOZYME | | Authors: | Vetter, I.R, Baase, W.A, Heinz, D.W, Xiong, J.-P, Snow, S, Matthews, B.W. | | Deposit date: | 1996-09-23 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Protein structural plasticity exemplified by insertion and deletion mutants in T4 lysozyme.
Protein Sci., 5, 1996
|
|
4MV0
 
 | | IspH in complex with pyridin-2-ylmethyl diphosphate | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER, pyridin-2-ylmethyl trihydrogen diphosphate | | Authors: | Span, I, Wang, K, Song, Y, Eisenreich, W, Bacher, A, Oldfield, E, Groll, M. | | Deposit date: | 2013-09-23 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the Binding of Pyridines to the Iron-Sulfur Enzyme IspH.
J.Am.Chem.Soc., 136, 2014
|
|
7LHK
 
 | |
1GXN
 
 | | Family 10 polysaccharide lyase from Cellvibrio cellulosa | | Descriptor: | PECTATE LYASE | | Authors: | Charnock, S.J, Brown, I.E, Turkenburg, J.P, Black, G.W, Davies, G.J. | | Deposit date: | 2002-04-08 | | Release date: | 2002-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Convergent Evolution Sheds Light on the Anti-Beta-Elimination Mechanism Common to Family 1 and 10 Polysaccharide Lyases
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3NVE
 
 | |
1GYZ
 
 | |
4MZI
 
 | | Crystal structure of a human mutant p53 | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Emamzadah, S, Tropia, L, Vincenti, I, Falquet, B, Halazonetis, T.D. | | Deposit date: | 2013-09-30 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Reversal of the DNA-Binding-Induced Loop L1 Conformational Switch in an Engineered Human p53 Protein.
J.Mol.Biol., 426, 2014
|
|
1NPD
 
 | | X-RAY STRUCTURE OF SHIKIMATE DEHYDROGENASE COMPLEXED WITH NAD+ FROM E.COLI (YDIB) NORTHEAST STRUCTURAL GENOMICS RESEARCH CONSORTIUM (NESG) TARGET ER24 | | Descriptor: | HYPOTHETICAL SHIKIMATE 5-DEHYDROGENASE-LIKE PROTEIN YDIB, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Benach, J, Kuzin, A.P, Lee, I, Rost, B, Chiang, Y, Acton, T.B, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-01-17 | | Release date: | 2003-01-28 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 2.3-A crystal structure of the shikimate 5-dehydrogenase orthologue YdiB from Escherichia coli suggests a novel catalytic environment for an NAD-dependent dehydrogenase
J.Biol.Chem., 278, 2003
|
|
4N1Q
 
 | |
2XQ5
 
 | | Pentameric ligand gated ion channel GLIC in complex with tetraethylarsonium (TEAs) | | Descriptor: | ARSENIC, GLR4197 PROTEIN | | Authors: | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
