1NSF
 
 | | D2 HEXAMERIZATION DOMAIN OF N-ETHYLMALEIMIDE SENSITIVE FACTOR (NSF) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, N-ETHYLMALEIMIDE SENSITIVE FACTOR | | Authors: | Yu, R.C, Hanson, P.I, Jahn, R, Brunger, A.T. | | Deposit date: | 1998-06-26 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the ATP-dependent oligomerization domain of N-ethylmaleimide sensitive factor complexed with ATP.
Nat.Struct.Biol., 5, 1998
|
|
4N1Z
 
 | | Crystal Structure of Human Farnesyl Diphosphate Synthase in Complex with BPH-1222 | | Descriptor: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Liu, Y.L, Xia, Y, Zhang, Y, Verma, I, Oldfield, E. | | Deposit date: | 2013-10-04 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A combination therapy for KRAS-driven lung adenocarcinomas using lipophilic bisphosphonates and rapamycin.
Sci Transl Med, 6, 2014
|
|
1NVN
 
 | | Structural Characterisation of the Holliday junction formed by the sequence CCGGTACCGG at 1.8 A | | Descriptor: | 5'-D(CpCpGpGpTpApCpCpGpG)-3', CALCIUM ION | | Authors: | Cardin, C.J, Gale, B.C, Thorpe, J.H, Teixeira, S.C.M, Gan, Y, Moraes, M.I.A.A, Brogden, A.L. | | Deposit date: | 2003-02-04 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of two Holliday junctions formed by the sequences TCGGTACCGA and CCGGTACCGG
To be Published
|
|
1NW5
 
 | | Structure of the beta class N6-adenine DNA methyltransferase RsrI bound to S-ADENOSYLMETHIONINE | | Descriptor: | CHLORIDE ION, MODIFICATION METHYLASE RSRI, S-ADENOSYLMETHIONINE | | Authors: | Thomas, C.B, Scavetta, R.D, Gumport, R.I, Churchill, M.E.A. | | Deposit date: | 2003-02-05 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of liganded and unliganded RsrI N6-adenine DNA methyltransferase: a distinct orientation for active cofactor binding
J.Biol.Chem., 278, 2003
|
|
1NXU
 
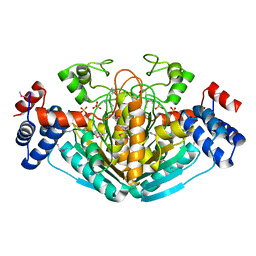 | | CRYSTAL STRUCTURE OF E. COLI HYPOTHETICAL OXIDOREDUCTASE YIAK NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ER82. | | Descriptor: | Hypothetical oxidoreductase yiaK, SULFATE ION | | Authors: | Forouhar, F, Lee, I, Benach, J, Kulkarni, K, Xiao, R, Acton, T.B, Shastry, R, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Novel NAD-binding Protein Revealed by the Crystal Structure of 2,3-Diketo-L-gulonate Reductase (YiaK).
J.Biol.Chem., 279, 2004
|
|
2CLI
 
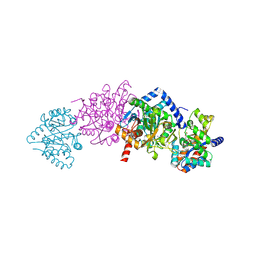 | | Tryptophan Synthase in complex with N-(4'- trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9) | | Descriptor: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Ngo, H, Harris, R, Kimmich, N, Casino, P, Niks, D, Blumenstein, L, Barends, T.R, Kulik, V, Weyand, M, Schlichting, I, Dunn, M.F. | | Deposit date: | 2006-04-27 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis and Characterization of Allosteric Probes of Substrate Channeling in the Tryptophan Synthase Bienzyme Complex.
Biochemistry, 46, 2007
|
|
4MQ0
 
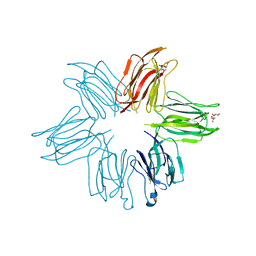 | | Crystal structure of Parkia biglobosa seed lectin (PBL) in complex with methyl alpha D-mannopyranoside | | Descriptor: | Parkia biglobosa lectin (PBL), methyl alpha-D-mannopyranoside | | Authors: | Teixeira, C.S, Rocha, B.A.M, Silva, H.C, Bari, A.U, Barroso-Neto, I.L, Santiago, M.Q, Nagano, C.S, Nascimento, K.S, Debray, H, Delatorre, P, Cavada, B.S. | | Deposit date: | 2013-09-14 | | Release date: | 2014-12-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of Parkia biglobosa seed lectin (PBL) in complex with methyl alpha D-mannopyranoside
To be Published
|
|
2CWI
 
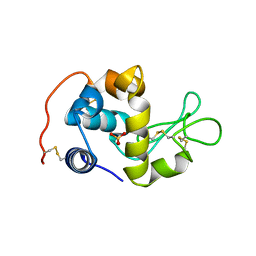 | | X-ray crystal structure analysis of recombinant wild-type canine milk lysozyme (apo-type) | | Descriptor: | Lysozyme C, milk isozyme, SULFATE ION | | Authors: | Akieda, D, Yasui, M, Aizawa, T, Yao, M, Watanabe, N, Tanaka, I, Demura, M, Kawano, K, Nitta, K. | | Deposit date: | 2005-06-20 | | Release date: | 2006-06-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Construction of an expression system of canine milk lysozyme in the methylotrophic yeast Pichia pastoris
To be Published
|
|
1NYX
 
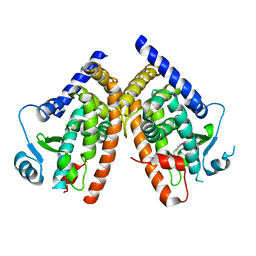 | | Ligand binding domain of the human peroxisome proliferator activated receptor gamma in complex with an agonist | | Descriptor: | (2S)-2-ETHOXY-3-{4-[2-(10H-PHENOXAZIN-10-YL)ETHOXY]PHENYL}PROPANOIC ACID, peroxisome proliferator activated receptor gamma | | Authors: | Ebdrup, S, Pettersson, I, Rasmussen, H.B, Deussen, H.-J, Frost Jensen, A, Mortensen, S.B, Fleckner, J, Pridal, L, Nygaard, L, Sauerberg, P. | | Deposit date: | 2003-02-14 | | Release date: | 2003-07-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthesis and biological and structural characterization of the dual-acting peroxisome proliferator-activated receptor alpha/gamma agonist ragaglitazar
J.MED.CHEM., 46, 2003
|
|
1O06
 
 | | Crystal structure of the Vps27p Ubiquitin Interacting Motif (UIM) | | Descriptor: | Vacuolar protein sorting-associated protein VPS27, ZINC ION | | Authors: | Fisher, R.D, Wang, B, Alam, S.L, Higginson, D.S, Rich, R, Myszka, D, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and ubiquitin binding of the ubiquitin-interacting motif.
J.Biol.Chem., 278, 2003
|
|
2CZ1
 
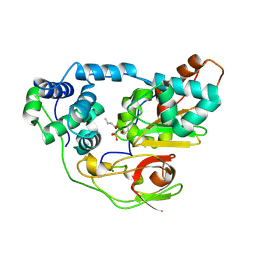 | | photo-activation state of Fe-type NHase with n-BA in anaerobic condition | | Descriptor: | BUTANOIC ACID, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Kawano, Y, Hashimoto, K, Odaka, M, Nakayama, H, Takio, K, Endo, I, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-09 | | Release date: | 2006-01-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | photo-activation state of Fe-type NHase with n-BA in anaerobic condition
To be Published
|
|
2CLH
 
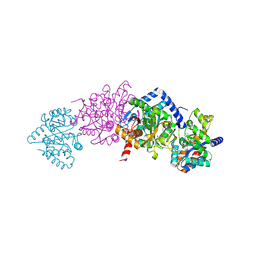 | | Tryptophan Synthase in complex with (naphthalene-2'-sulfonyl)-2-amino- 1-ethylphosphate (F19) | | Descriptor: | 2-[(2-NAPHTHYLSULFONYL)AMINO]ETHYL DIHYDROGEN PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Ngo, H, Harris, R, Kimmich, N, Casino, P, Niks, D, Blumenstein, L, Barends, T.R, Kulik, V, Weyand, M, Schlichting, I, Dunn, M.F. | | Deposit date: | 2006-04-27 | | Release date: | 2007-06-12 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis and characterization of allosteric probes of substrate channeling in the tryptophan synthase bienzyme complex.
Biochemistry, 46, 2007
|
|
1OHA
 
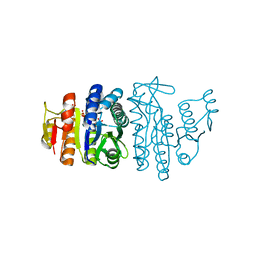 | | Acetylglutamate kinase from Escherichia coli complexed with MgADP and N-acetyl-L-glutamate | | Descriptor: | ACETATE ION, ACETYLGLUTAMATE KINASE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gil-Ortiz, F, Ramon-Maiques, S, Fita, I, Rubio, V. | | Deposit date: | 2003-05-23 | | Release date: | 2003-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Course of Phosphorus in the Reaction of N-Acetyl-L-Glutamate Kinase, Determined from the Structures of Crystalline Complexes, Including a Complex with an Alf(4)(-) Transition State Mimic
J.Mol.Biol., 331, 2003
|
|
1OMO
 
 | | alanine dehydrogenase dimer w/bound NAD (archaeal) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, alanine dehydrogenase | | Authors: | Gallagher, D.T, Smith, N.N, Holden, M.J, Schroeder, I, Monbouquette, H.G. | | Deposit date: | 2003-02-25 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of alanine dehydrogenase from Archaeoglobus: active site analysis and relation to bacterial cyclodeaminases and mammalian mu crystallin.
J.Mol.Biol., 342, 2004
|
|
3EFZ
 
 | | Crystal Structure of a 14-3-3 protein from cryptosporidium parvum (cgd1_2980) | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein | | Authors: | Wernimont, A.K, Dong, A, Qiu, W, Lew, J, Wasney, G.A, Vedadi, M, Kozieradzki, I, Zhao, Y, Ren, H, Alam, Z, Lin, Y.H, Sundstrom, M, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Hui, R, Brokx, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-10 | | Release date: | 2008-09-23 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Characterization of 14-3-3 proteins from Cryptosporidium parvum.
Plos One, 6, 2011
|
|
1Z6I
 
 | | Crystal structure of the ectodomain of Drosophila transmembrane receptor PGRP-LCa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Peptidoglycan-recognition protein-LC, SULFATE ION | | Authors: | Chang, C.-I, Ihara, K, Chelliah, Y, Mengin-Lecreulx, D, Wakatsuki, S, Deisenhofer, J. | | Deposit date: | 2005-03-22 | | Release date: | 2005-07-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the ectodomain of Drosophila peptidoglycan-recognition protein LCa suggests a molecular mechanism for pattern recognition
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZOR
 
 | | Isocitrate dehydrogenase from the hyperthermophile Thermotoga maritima | | Descriptor: | SODIUM ION, isocitrate dehydrogenase | | Authors: | Karlstrom, M, Steen, I.H, Birkeland, N.-K, Ladenstein, R. | | Deposit date: | 2005-05-13 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The crystal structure of a hyperthermostable subfamily II isocitrate dehydrogenase from Thermotoga maritima.
Febs J., 273, 2006
|
|
1OLQ
 
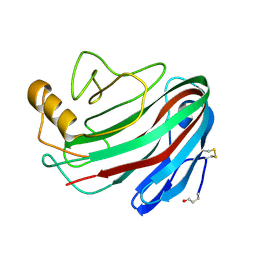 | | The Trichoderma reesei cel12a P201C mutant, structure at 1.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-BETA-1,4-GLUCANASE | | Authors: | Sandgren, M, Gualfetti, P.J, Shaw, A, Gross, L.S, Saldajeno, M, Berglund, G.I, Jones, T.A, Mitchinson, C. | | Deposit date: | 2003-08-11 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Humicola Grisea Cel12A Enzyme Structure at 1.2 A Resolution and the Impact of its Free Cysteine Residues on Thermal Stability
Protein Sci., 12, 2003
|
|
1OKB
 
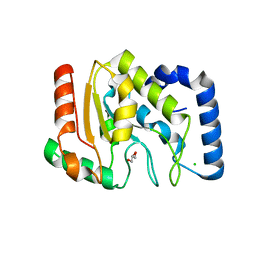 | | crystal structure of Uracil-DNA glycosylase from Atlantic cod (Gadus morhua) | | Descriptor: | CHLORIDE ION, GLYCEROL, URACIL-DNA GLYCOSYLASE | | Authors: | Leiros, I, Moe, E, Lanes, O, Smalas, A.O, Willassen, N.P. | | Deposit date: | 2003-07-21 | | Release date: | 2004-04-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Uracil-DNA Glycosylase from Atlantic Cod (Gadus Morhua) Reveals Cold-Adaptation Features
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1ZDU
 
 | | The Crystal Structure of Human Liver Receptor Homologue-1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nuclear receptor coactivator 2, Orphan nuclear receptor NR5A2, ... | | Authors: | Wang, W, Zhang, C, Marimuthu, A, Krupka, H.I, Tabrizizad, M, Shelloe, R, Mehra, U, Eng, K, Nguyen, H, Settachatgul, C, Powell, B, Milburn, M.V, West, B.L. | | Deposit date: | 2005-04-14 | | Release date: | 2005-05-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structures of human steroidogenic factor-1 and liver receptor homologue-1
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1OWX
 
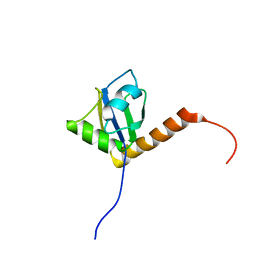 | | Solution structure of the C-terminal RRM of human La (La225-334) | | Descriptor: | Lupus La protein | | Authors: | Jacks, A, Babon, J, Kelly, G, Manolaridis, I, Cary, P.D, Curry, S, Conte, M.R. | | Deposit date: | 2003-03-31 | | Release date: | 2003-07-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal domain of human La protein reveals a novel RNA recognition motif coupled to a helical nuclear retention element
Structure, 11, 2003
|
|
1P9K
 
 | | THE SOLUTION STRUCTURE OF YBCJ FROM E. COLI REVEALS A RECENTLY DISCOVERED ALFAL MOTIF INVOLVED IN RNA-BINDING | | Descriptor: | orf, hypothetical protein | | Authors: | Volpon, L, Lievre, C, Osborne, M.J, Gandhi, S, Iannuzzi, P, Larocque, R, Matte, A, Cygler, M, Gehring, K, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-05-12 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of YbcJ from Escherichia coli reveals a recently discovered alphaL motif involved in RNA binding.
J.Bacteriol., 185, 2003
|
|
2AO6
 
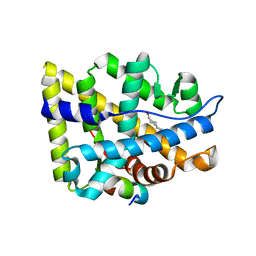 | | Crystal structure of the human androgen receptor ligand binding domain bound with TIF2(iii) 740-753 peptide and R1881 | | Descriptor: | (17BETA)-17-HYDROXY-17-METHYLESTRA-4,9,11-TRIEN-3-ONE, 14-mer fragment of Nuclear receptor coactivator 2, androgen receptor | | Authors: | He, B, Gampe Jr, R.T, Kole, A.J, Hnat, A.T, Stanley, T.B, An, G, Stewart, E.L, Kalman, R.I, Minges, J.T, Wilson, E.M. | | Deposit date: | 2005-08-12 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for androgen receptor interdomain and coactivator interactions suggests a transition in nuclear receptor activation function dominance
Mol.Cell, 16, 2004
|
|
2AWK
 
 | | GFP R96M mature chromophore | | Descriptor: | MAGNESIUM ION, green fluorescent protein | | Authors: | Wood, T.I, Barondeau, D.P, Hitomi, C, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2005-09-01 | | Release date: | 2006-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Defining the role of arginine 96 in green fluorescent protein fluorophore biosynthesis.
Biochemistry, 44, 2005
|
|
1OYE
 
 | | Structural Basis of Multiple Binding Capacity of the AcrB multidrug Efflux Pump | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, Acriflavine resistance protein B | | Authors: | Yu, E.W, McDermott, G, Zgurskaya, H.I, Nikaido, H, Koshland Jr, D.E. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump.
Science, 300, 2003
|
|
