5WEL
 
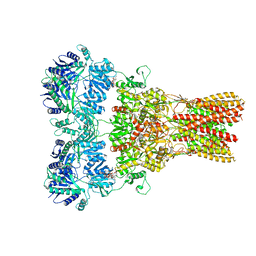 | | GluA2 bound to antagonist ZK and GSG1L in digitonin, state 2 | | Descriptor: | Chimera of Glutamate receptor 2, Germ cell-specific gene 1-like protein, Digitonin, ... | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2017-07-10 | | Release date: | 2017-08-02 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Channel opening and gating mechanism in AMPA-subtype glutamate receptors.
Nature, 549, 2017
|
|
1KVL
 
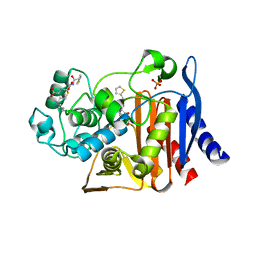 | | X-ray Crystal Structure of AmpC S64G Mutant beta-Lactamase in Complex with Substrate and Product Forms of Cephalothin | | Descriptor: | 2-[CARBOXY-(2-THIOPHEN-2-YL-ACETYLAMINO)-METHYL]-5-METHYL-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, 2-[CARBOXY-(2-THIOPHEN-2-YL-ACETYLAMINO)-METHYL]-5-METHYLENE-5,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Beta-lactamase, ... | | Authors: | Beadle, B.M, Trehan, I, Focia, P.J, Shoichet, B.K. | | Deposit date: | 2002-01-27 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural milestones in the reaction pathway of an amide hydrolase: substrate, acyl, and product complexes of cephalothin with AmpC beta-lactamase.
Structure, 10, 2002
|
|
1KWY
 
 | | Rat mannose protein A complexed with man-a13-man. | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN A, ... | | Authors: | Ng, K.K, Kolatkar, A.R, Park-Snyder, S, Feinberg, H, Clark, D.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2002-01-30 | | Release date: | 2002-07-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Orientation of bound ligands in mannose-binding proteins. Implications for multivalent ligand recognition.
J.Biol.Chem., 277, 2002
|
|
6CF7
 
 | |
4HHF
 
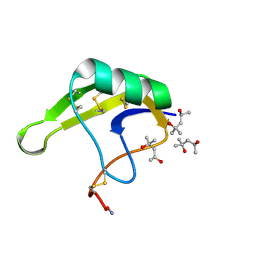 | | Crystal Structure of chemically synthesized scorpion alpha-toxin OD1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha-toxin OD1 | | Authors: | Wang, C.I.A, Lewis, R.J, Alewood, P.F, Durek, T. | | Deposit date: | 2012-10-09 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemical engineering and structural and pharmacological characterization of the alpha-scorpion toxin OD1.
Acs Chem.Biol., 8, 2013
|
|
5AIS
 
 | | Complex of human hematopoietic prostagandin D2 synthase (hH-PGDS) in complex with an active site inhibitor. | | Descriptor: | 4-(dimethylamino)-N-[5-(1H-indol-4-yl)pyridin-3-yl]butanamide, GLUTATHIONE, HEMATOPOIETIC PROSTAGLANDIN D SYNTHASE, ... | | Authors: | Edfeldt, F, Evenas, J, Lepisto, M, Ward, A, Petersen, J, Wissler, L, Rohman, M, Sivars, U, Svensson, K, Perry, M, Feierberg, I, Zhou, X, Hansson, T, Narjes, F. | | Deposit date: | 2015-02-17 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification of Indole Inhibitors of Human Hematopoietic Prostaglandin D2 Synthase (Hh-Pgds).
Bioorg.Med.Chem.Lett., 25, 2015
|
|
2W9B
 
 | | Binary complex of Dpo4 bound to N2,N2-dimethyl-deoxyguanosine modified DNA | | Descriptor: | 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*DOCP)-3', 5'-D(*TP*CP*AP*TP*M2GP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', DNA POLYMERASE IV, ... | | Authors: | Eoff, R.L, Zhang, H, Kosekov, I.D, Rizzo, C.J, Egli, M, Guengerich, F.P. | | Deposit date: | 2009-01-22 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure-Function Relationships in Miscoding by Sulfolobus Solfataricus DNA Polymerase Dpo4: Guanine N2,N2-Dimethyl Substitution Produces Inactive and Miscoding Polymerase Complexes.
J.Biol.Chem., 284, 2009
|
|
3MJ6
 
 | |
2W3X
 
 | | Crystal structure of a bifunctional hotdog fold thioesterase in enediyne biosynthesis, CalE7 | | Descriptor: | CALE7, GLYCEROL, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), ... | | Authors: | Kotaka, M, Kong, R, Qureshi, I, Ho, Q.S, Sun, H, Liew, C.W, Goh, L.P, Cheung, P, Mu, Y, Lescar, J, Liang, Z.X. | | Deposit date: | 2008-11-17 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Catalytic Mechanism of the Thioesterase Cale7 in Enediyne Biosynthesis.
J.Biol.Chem., 284, 2009
|
|
1KV5
 
 | | Structure of Trypanosoma brucei brucei TIM with the salt-bridge-forming residue Arg191 mutated to Ser | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-PHOSPHOGLYCOLIC ACID, GLYCEROL, ... | | Authors: | Kursula, I, Partanen, S, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-01-25 | | Release date: | 2002-03-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The importance of the conserved Arg191-Asp227 salt bridge of triosephosphate isomerase for folding, stability, and catalysis
FEBS Lett., 518, 2002
|
|
2W9C
 
 | | Ternary complex of Dpo4 bound to N2,N2-dimethyl-deoxyguanosine modified DNA with incoming dTTP | | Descriptor: | 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*DOCP)-3', 5'-D(*TP*CP*AP*CP*O2GP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', DNA POLYMERASE IV, ... | | Authors: | Eoff, R.L, Zhang, H, Kosekov, I.D, Rizzo, C.J, Egli, M, Guengerich, F.P. | | Deposit date: | 2009-01-22 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Function Relationships in Miscoding by Sulfolobus Solfataricus DNA Polymerase Dpo4: Guanine N2,N2-Dimethyl Substitution Produces Inactive and Miscoding Polymerase Complexes.
J.Biol.Chem., 284, 2009
|
|
1KWT
 
 | | Rat mannose binding protein A (native, MPD) | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN A | | Authors: | Ng, K.K.S, Kolatkar, A.R, Park-Snyder, S, Feinberg, H, Clark, D.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2002-01-30 | | Release date: | 2002-07-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Orientation of bound ligands in mannose-binding proteins. Implications for multivalent ligand recognition.
J.Biol.Chem., 277, 2002
|
|
5SYO
 
 | | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 3 nicotinic acetylcholine receptor in complex with Cytisine | | Descriptor: | (1R,5S)-1,2,3,4,5,6-HEXAHYDRO-8H-1,5-METHANOPYRIDO[1,2-A][1,5]DIAZOCIN-8-ONE, Soluble acetylcholine receptor, Neuronal acetylcholine receptor subunit alpha-3 chimera | | Authors: | Bobango, J, Wu, J, Talley, I.T, Talley, T.T. | | Deposit date: | 2016-08-11 | | Release date: | 2016-10-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 3 nicotinic acetylcholine receptor in complex with Cytisine
To Be Published
|
|
2W8F
 
 | | Aplysia californica AChBP bound to in silico compound 31 | | Descriptor: | (3-EXO)-3-(10,11-DIHYDRO-5H-DIBENZO[A,D][7]ANNULEN-5-YLOXY)-8,8-DIMETHYL-8-AZONIABICYCLO[3.2.1]OCTANE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Ulens, C, Akdemir, A, Jongejan, A, van Elk, R, Edink, E, Bertrand, S, Perrakis, A, Leurs, R, Smit, A.B, Sixma, T.K, Bertrand, D, de Esch, I.J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Use of Acetylcholine Binding Protein in the Search for Novel Alpha7 Nicotinic Receptor Ligands. In Silico Docking, Pharmacological Screening, and X- Ray Analysis.
J.Med.Chem., 52, 2009
|
|
1OFY
 
 | | three dimensional structure of the reduced form of nine-heme cytochrome c at ph 7.5 | | Descriptor: | ACETATE ION, GLYCEROL, HEME C, ... | | Authors: | Bento, I, Teixeira, V.H, Baptista, A.M, Soares, C.M, Matias, P.M, Carrondo, M.A. | | Deposit date: | 2003-04-22 | | Release date: | 2003-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redox-Bohr and Other Cooperativity Effects in the Nine-Heme Cytochrome C from Desulfovibrio Desulfuricans Atcc 27774: Crystallographic and Modeling Studies
J.Biol.Chem., 278, 2003
|
|
1KOZ
 
 | | SOLUTION STRUCTURE OF OMEGA-GRAMMOTOXIN SIA | | Descriptor: | Voltage-dependent Channel Inhibitor | | Authors: | Takeuchi, K, Park, E.J, Lee, C.W, Kim, J.I, Takahashi, H, Swartz, K.J, Shimada, I. | | Deposit date: | 2001-12-25 | | Release date: | 2002-08-28 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of omega-grammotoxin SIA, a gating modifier of P/Q and N-type Ca(2+) channel.
J.Mol.Biol., 321, 2002
|
|
5V68
 
 | | Crystal structure of cell division protein FtsZ from Mycobacterium tuberculosis bounded via the T9 loop | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Lazo, E.O, Ojima, I, Chowdhury, S.R, Awasthi, D, Jakoncic, J. | | Deposit date: | 2017-03-16 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Novel T9 loop conformation of filamenting temperature-sensitive mutant Z from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
1OFW
 
 | | Three dimensional structure of the oxidized form of nine heme cytochrome c at PH 7.5 | | Descriptor: | ACETATE ION, GLYCEROL, HEME C, ... | | Authors: | Bento, I, Teixeira, V.H, Baptista, A.M, Soares, C.M, Matias, P.M, Carrondo, M.A. | | Deposit date: | 2003-04-22 | | Release date: | 2003-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox-Bohr and Other Cooperativity Effects in the Nine-Heme Cytochrome C from Desulfovibrio Desulfuricans Atcc 27774: Crystallographic and Modeling Studies
J.Biol.Chem., 278, 2003
|
|
5T23
 
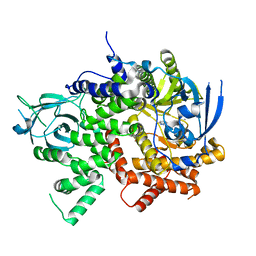 | | PI3Kg IN COMPLEX WITH 5d | | Descriptor: | 3-(benzotriazol-1-yl)-5-[1-methyl-5-[(1~{S})-1-morpholin-4-ylethyl]-1,2,4-triazol-3-yl]pyrazin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Petersen, J, Terstige, I, Perry, M, Svensson, T, Tyrchan, C, Lindmark, H, Oster, L. | | Deposit date: | 2016-08-23 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Discovery of triazole aminopyrazines as a highly potent and selective series of PI3K delta inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5T2B
 
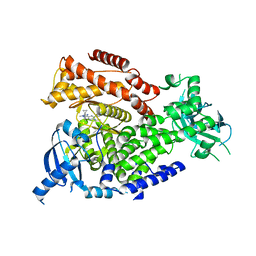 | | mPI3Kd IN COMPLEX WITH 5e | | Descriptor: | 5-[1-methyl-5-(1-phenylcyclopropyl)-1,2,4-triazol-3-yl]-3-(1~{H}-pyrazol-4-yl)pyrazin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Petersen, J, Terstige, I, Perry, M, Svensson, T, Tyrchan, C, Lindmark, H, Oster, L. | | Deposit date: | 2016-08-23 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of triazole aminopyrazines as a highly potent and selective series of PI3K delta inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5T2I
 
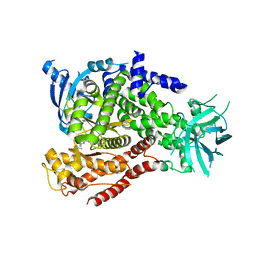 | | mPI3Kd IN COMPLEX WITH 7k | | Descriptor: | 1-[3-azanyl-6-[1-methyl-5-(1-phenylcyclopropyl)-1,2,4-triazol-3-yl]pyrazin-2-yl]pyrazole-4-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Petersen, J, Terstige, I, Perry, M, Svensson, T, Tyrchan, C, Lindmark, H, Oster, L. | | Deposit date: | 2016-08-23 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of triazole aminopyrazines as a highly potent and selective series of PI3K delta inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
1ONE
 
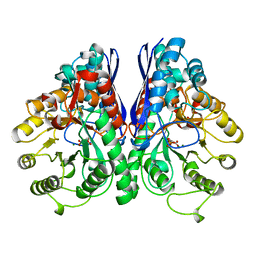 | | YEAST ENOLASE COMPLEXED WITH AN EQUILIBRIUM MIXTURE OF 2'-PHOSPHOGLYCEATE AND PHOSPHOENOLPYRUVATE | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, ENOLASE, MAGNESIUM ION, ... | | Authors: | Larsen, T.M, Wedekind, J.E, Rayment, I, Reed, G.H. | | Deposit date: | 1995-12-05 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A carboxylate oxygen of the substrate bridges the magnesium ions at the active site of enolase: structure of the yeast enzyme complexed with the equilibrium mixture of 2-phosphoglycerate and phosphoenolpyruvate at 1.8 A resolution.
Biochemistry, 35, 1996
|
|
1OTG
 
 | | 5-CARBOXYMETHYL-2-HYDROXYMUCONATE ISOMERASE | | Descriptor: | 5-CARBOXYMETHYL-2-HYDROXYMUCONATE ISOMERASE, SULFATE ION | | Authors: | Subramanya, H.S, Roper, D.I, Dauter, Z, Dodson, E.J, Davies, G.J, Wilson, K.S, Wigley, D.B. | | Deposit date: | 1995-11-09 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enzymatic ketonization of 2-hydroxymuconate: specificity and mechanism investigated by the crystal structures of two isomerases.
Biochemistry, 35, 1996
|
|
1KCF
 
 | | Crystal Structure of the Yeast Mitochondrial Holliday Junction Resolvase, Ydc2 | | Descriptor: | HYPOTHETICAL 30.2 KD PROTEIN C25G10.02 IN CHROMOSOME I, SULFATE ION | | Authors: | Ceschini, S, Keeley, A, McAlister, M.S.B, Oram, M, Phelan, J, Pearl, L.H, Tsaneva, I.R, Barrett, T.E. | | Deposit date: | 2001-11-08 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the fission yeast mitochondrial Holliday junction resolvase Ydc2.
EMBO J., 20, 2001
|
|
5T2D
 
 | | mPI3Kd IN COMPLEX WITH 7j | | Descriptor: | 4-[3-azanyl-6-[1-methyl-5-(1-phenylcyclopropyl)-1,2,4-triazol-3-yl]pyrazin-2-yl]-~{N},~{N}-dimethyl-pyrazole-1-sulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Petersen, J, Terstige, I, Perry, M, Svensson, T, Tyrchan, C, Lindmark, H, Oster, L. | | Deposit date: | 2016-08-23 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of triazole aminopyrazines as a highly potent and selective series of PI3K delta inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
