1QEY
 
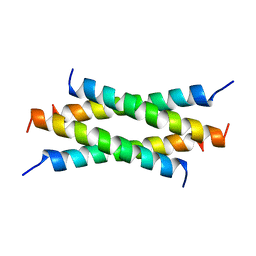 | | NMR Structure Determination of the Tetramerization Domain of the MNT Repressor: An Asymmetric A-Helical Assembly in Slow Exchange | | Descriptor: | PROTEIN (REGULATORY PROTEIN MNT) | | Authors: | Nooren, I.M.A, George, A.V.E, Kaptein, R, Sauer, R.T, Boelens, R. | | Deposit date: | 1999-04-03 | | Release date: | 1999-08-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The tetramerization domain of the Mnt repressor consists of two right-handed coiled coils.
Nat.Struct.Biol., 6, 1999
|
|
8PWR
 
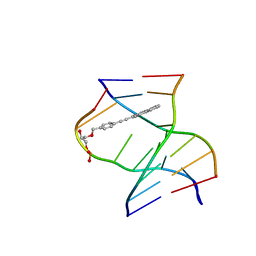 | | TINA-conjugated antiparallel DNA triplex | | Descriptor: | DNA (5'-D(*AP*GP*GP*AP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*TP*CP*CP*T)-3'), DNA (5'-D(*TP*GP*GP*TP*GP*(J32)P*GP*T)-3') | | Authors: | Garavis, M, Edwards, P.J.B, Serrano-Chacon, I, Doluca, O, Filichev, V.V, Gonzalez, C. | | Deposit date: | 2023-07-21 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-27 | | Method: | SOLUTION NMR | | Cite: | Understanding intercalative modulation of G-rich sequence folding: solution structure of a TINA-conjugated antiparallel DNA triplex.
Nucleic Acids Res., 52, 2024
|
|
6E2G
 
 | |
2VF2
 
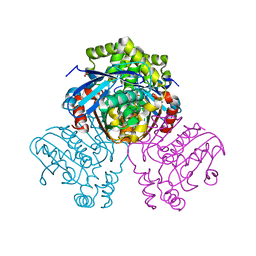 | | X-ray crystal structure of HsaD from Mycobacterium tuberculosis | | Descriptor: | 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, GLYCEROL, SULFATE ION | | Authors: | Lack, N, Lowe, E.D, Liu, J, Eltis, L.D, Noble, M.E.M, Sim, E, Westwood, I.M. | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Hsad, a Steroid-Degrading Hydrolase, from Mycobacterium Tuberculosis.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
5F5W
 
 | |
2N0M
 
 | | The solution structure of the soluble form of the Lipid-modified Azurin from Neisseria gonorrhoeae | | Descriptor: | COPPER (I) ION, Lipid modified azurin protein | | Authors: | Pauleta, S.R, Matzapetakis, M.F, Nobrega, C.F, Carreira, C, Saraiva, I.H. | | Deposit date: | 2015-03-10 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the soluble form of the lipid-modified azurin from Neisseria gonorrhoeae, the electron donor of cytochrome c peroxidase.
Biochim.Biophys.Acta, 1857, 2016
|
|
1KCF
 
 | | Crystal Structure of the Yeast Mitochondrial Holliday Junction Resolvase, Ydc2 | | Descriptor: | HYPOTHETICAL 30.2 KD PROTEIN C25G10.02 IN CHROMOSOME I, SULFATE ION | | Authors: | Ceschini, S, Keeley, A, McAlister, M.S.B, Oram, M, Phelan, J, Pearl, L.H, Tsaneva, I.R, Barrett, T.E. | | Deposit date: | 2001-11-08 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the fission yeast mitochondrial Holliday junction resolvase Ydc2.
EMBO J., 20, 2001
|
|
6E2F
 
 | |
1KAJ
 
 | |
6ELX
 
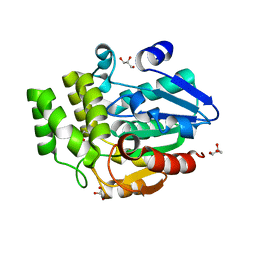 | | Oryza sativa DWARF14 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, ... | | Authors: | Andersson, I, Carlsson, G.H, Hasse, D. | | Deposit date: | 2017-09-29 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The elusive ligand complexes of the DWARF14 strigolactone receptor.
J. Exp. Bot., 69, 2018
|
|
1KH3
 
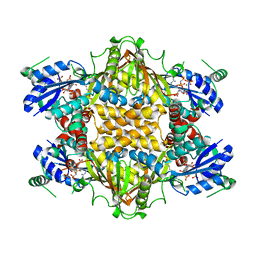 | | Crystal Structure of Thermus thermophilus HB8 Argininosuccinate Synthetase in complex with inhibitor | | Descriptor: | ARGININE, ASPARTIC ACID, Argininosuccinate Synthetase, ... | | Authors: | goto, m, Hirotsu, k, miyahara, i, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-11-29 | | Release date: | 2003-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Argininosuccinate Synthetase in Enzyme-ATP Substrates and Enzyme-AMP Product Forms: STEREOCHEMISTRY OF THE CATALYTIC REACTION
J.Biol.Chem., 278, 2003
|
|
6E3E
 
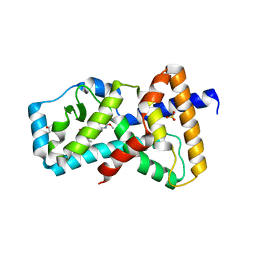 | | Structure of RORgt in complex with a novel inverse agonist. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-[(5R)-5-[(7-fluoro-1,1-dimethyl-2,3-dihydro-1H-inden-5-yl)carbamoyl]-2-methoxy-7,8-dihydro-1,6-naphthyridin-6(5H)-yl]-5-oxopentanoic acid, Nuclear receptor ROR-gamma, ... | | Authors: | Skene, R.J, Hoffman, I. | | Deposit date: | 2018-07-13 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Retinoic Acid-Related Orphan Receptor gamma t (ROR gamma t) Agonist Structure-Based Functionality Switching Approach from In House ROR gamma t Inverse Agonist to ROR gamma t Agonist.
J.Med.Chem., 62, 2019
|
|
5B49
 
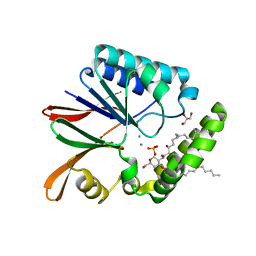 | | Crystal structure of LpxH with manganese from Pseudomonas aeruginosa | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Okada, C, Wakabayashi, H, Yao, M, Tanaka, I. | | Deposit date: | 2016-04-03 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of the UDP-diacylglucosamine pyrophosphohydrase LpxH from Pseudomonas aeruginosa
Sci Rep, 6, 2016
|
|
6YE5
 
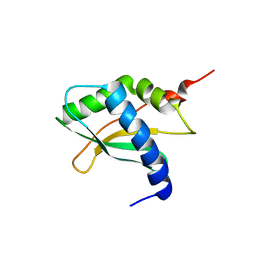 | | Structure of ribosomal binding factor A RbfA of Staphylococcus aureus bacterium by NMR | | Descriptor: | Ribosome-binding factor A | | Authors: | Blokhin, D.S, Usachev, K.S, Bikmullin, A.G, Nurullina, L, Garaeva, N, Validov, S, Klochkov, V, Aganov, A, Khusainov, I, Yusupov, M. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of ribosomal binding factor A RbfA of Staphylococcus aureus bacterium by NMR
To Be Published
|
|
6RF7
 
 | | Crystal structure of the light-driven sodium pump KR2 in the monomeric form, pH 8.9 | | Descriptor: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
8A9E
 
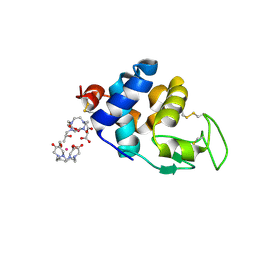 | | Lysozyme, 9-11 fs FEL pulses as determined by XTCAV | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, GADOLINIUM ATOM, Lysozyme | | Authors: | Barends, T, Nass, K, Gorel, A, Schlichting, I. | | Deposit date: | 2022-06-28 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Microcrystallization methods
To Be Published
|
|
8PAB
 
 | |
6E3G
 
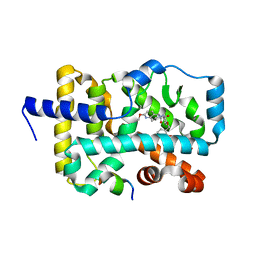 | | Structure of RORgt in complex with a novel agonist. | | Descriptor: | (5R)-6-acetyl-2-methoxy-N-{4-[(2-methoxyphenyl)methoxy]phenyl}-5,6,7,8-tetrahydro-1,6-naphthyridine-5-carboxamide, 1,2-ETHANEDIOL, Nuclear receptor ROR-gamma, ... | | Authors: | Skene, R.J, Hoffman, I. | | Deposit date: | 2018-07-13 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Retinoic Acid-Related Orphan Receptor gamma t (ROR gamma t) Agonist Structure-Based Functionality Switching Approach from In House ROR gamma t Inverse Agonist to ROR gamma t Agonist.
J.Med.Chem., 62, 2019
|
|
5B70
 
 | | OxyR2 E204G regulatory domain from Vibrio vulnificus | | Descriptor: | GLYCEROL, LysR family transcriptional regulator | | Authors: | Jo, I, Ha, N.-C. | | Deposit date: | 2016-06-02 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The hydrogen peroxide hypersensitivity of OxyR2 in Vibrio vulnificus depends on conformational constraints
J. Biol. Chem., 292, 2017
|
|
5T2L
 
 | | mPI3Kd IN COMPLEX WITH 7l | | Descriptor: | 1-[3-azanyl-6-[1-methyl-5-(1-phenylcyclopropyl)-1,2,4-triazol-3-yl]pyrazin-2-yl]pyrazole-3-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Petersen, J, Terstige, I, Perry, M, Svensson, T, Tyrchan, C, Lindmark, H, Oster, L. | | Deposit date: | 2016-08-23 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of triazole aminopyrazines as a highly potent and selective series of PI3K delta inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6TZB
 
 | |
6E3H
 
 | | Crystal structure of S9-3-37 bound to H5 influenza hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Wu, N.C, Wilson, I.A. | | Deposit date: | 2018-07-14 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The D3-9 gene segment encodes for recurring and adaptable binding motifs in broadly neutralizing antibodies to influenza virus
Cell Host Microbe, 2018
|
|
2DSO
 
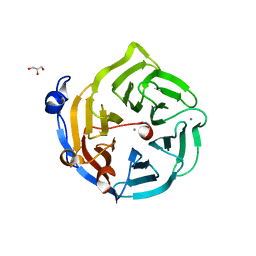 | | Crystal structure of D138N mutant of Drp35, a 35kDa drug responsive protein from Staphylococcus aureus | | Descriptor: | CALCIUM ION, Drp35, GLYCEROL | | Authors: | Tanaka, Y, Ohki, Y, Morikawa, K, Yao, M, Watanabe, N, Ohta, T, Tanaka, I. | | Deposit date: | 2006-07-04 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mutational Analyses of Drp35 from Staphylococcus aureus: A POSSIBLE MECHANISM FOR ITS LACTONASE ACTIVITY
J.Biol.Chem., 282, 2007
|
|
1KQV
 
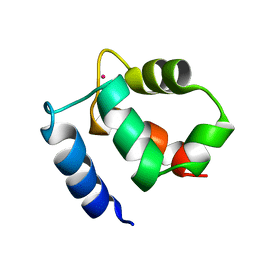 | | Family of NMR Solution Structures of Ca Ln Calbindin D9K | | Descriptor: | LANTHANUM (III) ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN | | Authors: | Bertini, I, Donaire, A, Jimenez, B, Luchinat, C, Parigi, G, Piccioli, M, Poggi, L. | | Deposit date: | 2002-01-08 | | Release date: | 2002-01-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Paramagnetism-based versus classical constraints: an analysis of the solution structure of Ca Ln calbindin D9k.
J.Biomol.NMR, 21, 2001
|
|
8AHU
 
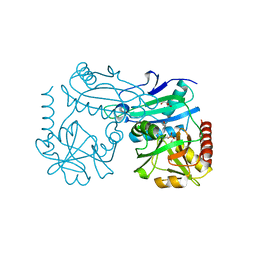 | | Crystal structure of D-amino acid aminotrensferase from Haliscomenobacter hydrossis complexed with D-cycloserine | | Descriptor: | Aminotransferase class IV, GLYCEROL, [5-hydroxy-6-methyl-4-({[(4E)-3-oxo-1,2-oxazolidin-4-ylidene]amino}methyl)pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Bakunova, A.K, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-07-22 | | Release date: | 2022-08-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Mechanism of D-Cycloserine Inhibition of D-Amino Acid Transaminase from Haliscomenobacter hydrossis.
Biochemistry Mosc., 88, 2023
|
|
