6XC4
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 heavy chain, CC12.3 light chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
1A8H
 
 | | METHIONYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS | | Descriptor: | METHIONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Sugiura, I, Nureki, O, Ugaji, Y, Kuwabara, S, Lober, B, Giege, R, Moras, D, Yokoyama, S, Konno, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-03-26 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A crystal structure of Thermus thermophilus methionyl-tRNA synthetase reveals two RNA-binding modules.
Structure, 8, 2000
|
|
7UFI
 
 | | VchTnsC AAA+ ATPase with DNA, single heptamer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*CP*TP*CP*CP*AP*GP*TP*AP*CP*AP*GP*CP*GP*CP*GP*GP*CP*TP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*CP*AP*GP*CP*CP*GP*CP*GP*CP*TP*GP*TP*AP*CP*TP*GP*GP*AP*G)-3'), ... | | Authors: | Fernandez, I.S, Sternberg, S.H. | | Deposit date: | 2022-03-22 | | Release date: | 2022-06-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Selective TnsC recruitment enhances the fidelity of RNA-guided transposition.
Nature, 609, 2022
|
|
5L8I
 
 | | crystal structure of human FABP6 apo-protein | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, DI(HYDROXYETHYL)ETHER, Gastrotropin, ... | | Authors: | Hendrick, A, Mueller, I, Leonard, P.M, Davenport, R, Mitchell, P. | | Deposit date: | 2016-06-08 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Identification and Investigation of Novel Binding Fragments in the Fatty Acid Binding Protein 6 (FABP6).
J.Med.Chem., 59, 2016
|
|
8ELQ
 
 | |
5L8N
 
 | | crystal structure of human FABP6 protein with fragment 1 | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 5,6-dimethyl-1~{H}-benzimidazol-2-amine, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hendrick, A, Mueller, I, Leonard, P.M, Davenport, R, Mitchell, P. | | Deposit date: | 2016-06-08 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Identification and Investigation of Novel Binding Fragments in the Fatty Acid Binding Protein 6 (FABP6).
J.Med.Chem., 59, 2016
|
|
5AY4
 
 | | Crystal structure of RNA duplex containing C-C base pairs obtained in the presence of Hg(II) | | Descriptor: | RNA (5'-R(*GP*GP*AP*CP*UP*(CBR)P*GP*AP*CP*UP*CP*C)-3'), SODIUM ION | | Authors: | Kondo, J, Tada, Y, Dairaku, T, Saneyoshi, H, Okamoto, I, Tanaka, Y, Ono, A. | | Deposit date: | 2015-08-06 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-Resolution Crystal Structure of a Silver(I)-RNA Hybrid Duplex Containing Watson-Crick-like CSilver(I)C Metallo-Base Pairs
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6YUE
 
 | | Fragment of nitrate/nitrite sensor histidine kinase NarQ (R50S variant) | | Descriptor: | Nitrate/nitrite sensor protein NarQ | | Authors: | Gushchin, I, Melnikov, I, Polovinkin, V, Yuzhakova, A, Gordeliy, V. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sensor Histidine Kinase NarQ Activates via Helical Rotation, Diagonal Scissoring, and Eventually Piston-Like Shifts.
Int J Mol Sci, 21, 2020
|
|
6YUL
 
 | | CK2 alpha bound to Macrocycle | | Descriptor: | 7,10-Dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.12,6.017,20]docosa-1(20),2(22),3,5,14(21),15,18-heptaene-5-carboxylic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Kraemer, A, Hanke, T, Kurz, C, Celik, I, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-27 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of pyrazolo[1,5-a]pyrimidines lead to the identification of a highly selective casein kinase 2 inhibitor.
Eur.J.Med.Chem., 208, 2020
|
|
4U8L
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant N207A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
8OR3
 
 | | CAND1-CUL1-RBX1-SKP1-SKP2-DCNL1 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, DCN1-like protein 1, ... | | Authors: | Shaaban, M, Clapperton, J.A, Ding, S, Maeots, M.E, Enchev, R.I. | | Deposit date: | 2023-04-13 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange.
Mol.Cell, 83, 2023
|
|
8OR0
 
 | | CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, Cyclin-dependent kinase 2, ... | | Authors: | Shaaban, M, Clapperton, J.A, Ding, S, Maeots, M.E, Enchev, R.I. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange.
Mol.Cell, 83, 2023
|
|
2RNB
 
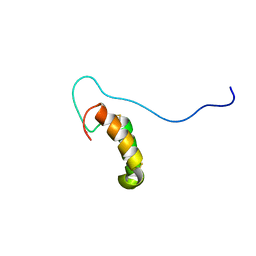 | | Solution structure of human Cu(I)Cox17 | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase copper chaperone | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Janicka, A, Martinelli, M, Kozlowski, H, Palumaa, P. | | Deposit date: | 2007-12-08 | | Release date: | 2007-12-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A structural-dynamical characterization of human cox17
J.Biol.Chem., 283, 2008
|
|
7BVR
 
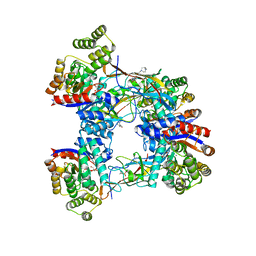 | | DgpB-DgpC complex apo | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AP_endonuc_2 domain-containing protein, DgpB, ... | | Authors: | Mori, T, He, H, Abe, I. | | Deposit date: | 2020-04-11 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
5IZD
 
 | |
1A1X
 
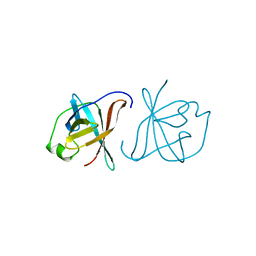 | | CRYSTAL STRUCTURE OF MTCP-1 INVOLVED IN T CELL MALIGNANCIES | | Descriptor: | HMTCP-1 | | Authors: | Fu, Z.Q, Dubois, G.C, Song, S.P, Kulikovskaya, I, Virgilio, L, Rothstein, J, Croce, C.M, Weber, I.T, Harrison, R.W. | | Deposit date: | 1997-12-18 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of MTCP-1: implications for role of TCL-1 and MTCP-1 in T cell malignancies.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1A5Z
 
 | | LACTATE DEHYDROGENASE FROM THERMOTOGA MARITIMA (TMLDH) | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CADMIUM ION, L-LACTATE DEHYDROGENASE, ... | | Authors: | Auerbach, G, Ostendorp, R, Prade, L, Korndoerfer, I, Dams, T, Huber, R, Jaenicke, R. | | Deposit date: | 1998-02-18 | | Release date: | 1999-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lactate dehydrogenase from the hyperthermophilic bacterium thermotoga maritima: the crystal structure at 2.1 A resolution reveals strategies for intrinsic protein stabilization.
Structure, 6, 1998
|
|
4U8N
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant F66A complexed with UDP | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
5J3N
 
 | | C-terminal domain of EcoR124I HsdR subunit fused with the pH-sensitive GFP variant ratiometric pHluorin | | Descriptor: | Green fluorescent protein,HsdR | | Authors: | Grinkevich, P, Iermak, I, Luedtke, N, Mesters, J.R, Ettrich, R, Ludwig, J. | | Deposit date: | 2016-03-31 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of a novel domain of the motor subunit of the Type I restriction enzyme EcoR124 involved in complex assembly and DNA binding.
J. Biol. Chem., 293, 2018
|
|
6YUM
 
 | | CK2 alpha bound to unclosed Macrocycle | | Descriptor: | 4-[5-[2-(2-hydroxyethyloxy)ethyl-[(2-methylpropan-2-yl)oxycarbonyl]amino]pyrazolo[1,5-a]pyrimidin-3-yl]-2-oxidanyl-benzoic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Kraemer, A, Hanke, T, Kurz, C, Celik, I, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-27 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Optimization of pyrazolo[1,5-a]pyrimidines lead to the identification of a highly selective casein kinase 2 inhibitor.
Eur.J.Med.Chem., 208, 2020
|
|
7W1D
 
 | | Crystal structure of Klebsiella pneumoniae K1 capsule-specific polysaccharide lyase in a C2 crystal form | | Descriptor: | CARBONATE ION, CITRIC ACID, K1 LYASE | | Authors: | Tu, I.F, Ko, T.P, Huang, K.F, Wu, S.H. | | Deposit date: | 2021-11-19 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural and biological insights into Klebsiella pneumoniae surface polysaccharide degradation by a bacteriophage K1 lyase: implications for clinical use.
J.Biomed.Sci., 29, 2022
|
|
6CHT
 
 | | HNF4alpha in complex with the corepressor EBP1 fragment | | Descriptor: | Hepatocyte nuclear factor 4-alpha, LAURIC ACID, Proliferation-associated protein 2G4 | | Authors: | Chi, Y.I, Singh, P, Lee, I.K. | | Deposit date: | 2018-02-22 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.174 Å) | | Cite: | ErbB3-binding protein 1 (EBP1) represses HNF4 alpha-mediated transcription and insulin secretion in pancreatic beta-cells.
J.Biol.Chem., 294, 2019
|
|
4U8K
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Q107A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8J
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Y104A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8M
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Y317A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
