2HWY
 
 | |
2HW0
 
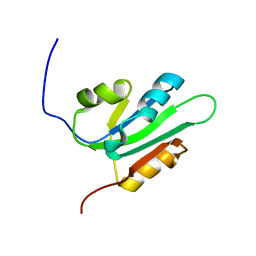 | | NMR Solution Structure of the nuclease domain from the Replicator Initiator Protein from porcine circovirus PCV2 | | Descriptor: | Replicase | | Authors: | Vega-Rocha, S, Byeon, I.L, Gronenborn, B, Gronenborn, A.M, Campos-Olivas, R. | | Deposit date: | 2006-07-31 | | Release date: | 2007-06-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure, divalent metal and DNA binding of the endonuclease domain from the replication initiation protein from porcine circovirus 2
J.Mol.Biol., 367, 2007
|
|
2K90
 
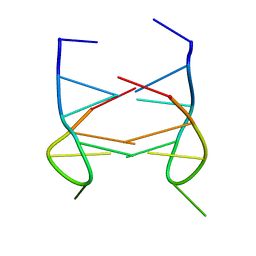 | | Dimeric solution structure of the DNA loop d(TGCTTCGT) | | Descriptor: | 5'-D(*TP*GP*CP*TP*TP*CP*GP*T)-3' | | Authors: | Viladoms, J, Escaja, N, Frieden, M, Gomez-Pinto, I, Pedroso, E, Gonzalez, C. | | Deposit date: | 2008-09-29 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Self-association of short DNA loops through minor groove C:G:G:C tetrads.
Nucleic Acids Res., 37, 2009
|
|
2KVR
 
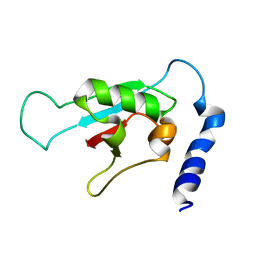 | | Solution NMR structure of human ubiquitin specific protease Usp7 UBL domain (residues 537-664). NESG target hr4395c/ SGC-Toronto | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Bezsonova, I, Lemak, A, Avvakumov, G, Xue, S, Dhe-Paganon, S, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-25 | | Release date: | 2010-04-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | solution structure of the ubiquitin specific protease Usp7 ubiquitin-like domain
To be Published
|
|
1DTO
 
 | | CRYSTAL STRUCTURE OF THE COMPLETE TRANSACTIVATION DOMAIN OF E2 PROTEIN FROM THE HUMAN PAPILLOMAVIRUS TYPE 16 | | Descriptor: | REGULATORY PROTEIN E2 | | Authors: | Antson, A.A, Burns, J.E, Moroz, O.V, Scott, D.J, Sanders, C.M, Bronstein, I.B, Dodson, G.G, Wilson, K.S, Maitland, N. | | Deposit date: | 2000-01-13 | | Release date: | 2000-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the intact transactivation domain of the human papillomavirus E2 protein.
Nature, 403, 2000
|
|
1DQD
 
 | | CRYSTAL STRUCTURE OF FAB HGR-2 F6, A COMPETITIVE ANTAGONIST OF THE GLUCAGON RECEPTOR | | Descriptor: | FAB HGR-2 F6 | | Authors: | Wright, L.M, Brzozowski, A.M, Hubbard, R.E, Pike, A.C.W, Roberts, S.M, Skovgaard, R.N, Svendsen, I, Vissing, H, Bywater, R.P. | | Deposit date: | 2000-01-04 | | Release date: | 2000-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Fab hGR-2 F6, a competitive antagonist of the glucagon receptor.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1DYQ
 
 | | STAPHYLOCOCCAL ENTEROTOXIN A MUTANT VACCINE | | Descriptor: | Enterotoxin type A, SULFATE ION, ZINC ION | | Authors: | Krupka, H.I, Segelke, B.W, Rupp, B. | | Deposit date: | 2000-02-05 | | Release date: | 2001-03-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Abrogated Binding between Staphylococcal Enterotoxin a Superantigen Vaccine and Mhc-II?
Protein Sci., 11, 2002
|
|
2K97
 
 | | Dimeric solution structure of the cyclic octamer d(pCGCTCCGT) | | Descriptor: | 5'-D(P*CP*GP*CP*TP*CP*CP*GP*T)-3' | | Authors: | Viladoms, J, Escaja, N, Frieden, M, Gomez-Pinto, I, Pedroso, E, Gonzalez, C. | | Deposit date: | 2008-09-30 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Self-association of short DNA loops through minor groove C:G:G:C tetrads.
Nucleic Acids Res., 37, 2009
|
|
2K9Z
 
 | | NMR structure of the protein TM1112 | | Descriptor: | uncharacterized protein TM1112 | | Authors: | Mohanty, B, Pedrini, B, Serrano, P, Geralt, M, Horst, R, Herrmann, T, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-28 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures for the proteins TM1112 and TM1367.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1DFS
 
 | | SOLUTION STRUCTURE OF THE ALPHA-DOMAIN OF MOUSE METALLOTHIONEIN-1 | | Descriptor: | CADMIUM ION, METALLOTHIONEIN-1 | | Authors: | Zangger, K, Oz, G, Otvos, J.D, Armitage, I.M. | | Deposit date: | 1999-11-20 | | Release date: | 1999-12-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of mouse [Cd7]-metallothionein-1 by homonuclear and heteronuclear NMR spectroscopy.
Protein Sci., 8, 1999
|
|
2L84
 
 | | Solution NMR structures of CBP bromodomain with small molecule j28 | | Descriptor: | 5-[(E)-(2-amino-4-hydroxy-5-methylphenyl)diazenyl]-2,4-dimethylbenzenesulfonic acid, CREB-binding protein | | Authors: | Borah, J.C, Mujtaba, S, Karakikes, I, Zeng, L, Muller, M, Patel, J, Moshkina, N, Morohashi, K, Zhang, W, Gerona-Navarro, G, Hajjar, R.J, Zhou, M. | | Deposit date: | 2011-01-03 | | Release date: | 2011-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule Binding to the Coactivator CREB-Binding Protein Blocks Apoptosis in Cardiomyocytes.
Chem.Biol., 18, 2011
|
|
2L1O
 
 | | Zinc to cadmium replacement in the A. thaliana SUPERMAN Cys2His2 zinc finger induces structural rearrangements of typical DNA base determinant positions | | Descriptor: | CADMIUM ION, Transcriptional regulator SUPERMAN | | Authors: | Malgieri, G, Zaccaro, L, Leone, M, Bucci, E, Esposito, S, Baglivo, I, Del Gatto, A, Scandurra, R, Pedone, P.V, Fattorusso, R, Isernia, C. | | Deposit date: | 2010-07-31 | | Release date: | 2011-06-08 | | Last modified: | 2011-12-21 | | Method: | SOLUTION NMR | | Cite: | Zinc to cadmium replacement in the A. thaliana SUPERMAN Cys(2) His(2) zinc finger induces structural rearrangements of typical DNA base determinant positions.
Biopolymers, 95, 2011
|
|
1D9W
 
 | |
1DCS
 
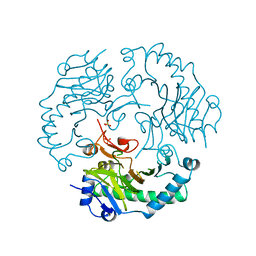 | | DEACETOXYCEPHALOSPORIN C SYNTHASE FROM S. CLAVULIGERUS | | Descriptor: | DEACETOXYCEPHALOSPORIN C SYNTHASE, SULFATE ION | | Authors: | Valegard, K, Terwisscha Van Scheltinga, A.C, Lloyd, M.D, Hara, T, Ramaswamy, S, Perrakis, A, Thompson, A, Lee, H.J, Baldwin, J.E, Schofield, C.J, Hajdu, J, Andersson, I. | | Deposit date: | 1998-06-05 | | Release date: | 1999-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of a cephalosporin synthase.
Nature, 394, 1998
|
|
2FPF
 
 | |
2FPL
 
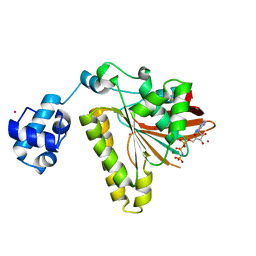 | | RadA recombinase in complex with AMP-PNP and low concentration of K+ | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Wu, Y, Qian, X, He, Y, Moya, I.A, Luo, Y. | | Deposit date: | 2006-01-16 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Methanococcus Voltae Rada in Complex with Adp: hydrolysis-induced conformational change
Biochemistry, 44, 2005
|
|
2DQJ
 
 | | Crystal structure of hyhel-10 FV (wild-type) complexed with hen egg lysozyme at 1.8A resolution | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Shiroishi, M, Kondo, H, Tsumoto, K, Kumagai, I. | | Deposit date: | 2006-05-26 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural consequences of mutations in interfacial Tyr residues of a protein antigen-antibody complex. The case of HyHEL-10-HEL
J.Biol.Chem., 282, 2007
|
|
2LD4
 
 | | Solution structure of the N-terminal domain of human anamorsin | | Descriptor: | Anamorsin | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Boscaro, F, Chatzi, A, Mikolajczyk, M, Tokatlidis, K, Winkelmann, J. | | Deposit date: | 2011-05-13 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Anamorsin Is a [2Fe-2S] Cluster-Containing Substrate of the Mia40-Dependent Mitochondrial Protein Trapping Machinery.
Chem.Biol., 18, 2011
|
|
2L51
 
 | | Solution structure of calcium bound S100A16 | | Descriptor: | CALCIUM ION, Protein S100-A16 | | Authors: | Babini, E, Bertini, I, Borsi, V, Calderone, V, Hu, X, Luchinat, C, Parigi, G. | | Deposit date: | 2010-10-22 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of human S100A16, a low-affinity calcium binder.
J.Biol.Inorg.Chem., 16, 2011
|
|
2LLT
 
 | | Post-translational S-nitrosylation is an endogenous factor fine-tuning human S100A1 protein properties | | Descriptor: | Protein S100-A1 | | Authors: | Lenarcic Zivkovic, M, Zareba-Koziol, M, Zhukova, L, Poznanski, J, Zhukov, I, Wyslouch-Cieszynska, A. | | Deposit date: | 2011-11-17 | | Release date: | 2012-09-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Post-translational S-Nitrosylation Is an Endogenous Factor Fine Tuning the Properties of Human S100A1 Protein.
J.Biol.Chem., 287, 2012
|
|
1DFT
 
 | | SOLUTION STRUCTURE OF THE BETA-DOMAIN OF MOUSE METALLOTHIONEIN-1 | | Descriptor: | CADMIUM ION, METALLOTHIONEIN-1 | | Authors: | Zangger, K, Oz, G, Otvos, J.D, Armitage, I.M. | | Deposit date: | 1999-11-20 | | Release date: | 1999-12-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of mouse [Cd7]-metallothionein-1 by homonuclear and heteronuclear NMR spectroscopy.
Protein Sci., 8, 1999
|
|
2G5H
 
 | | Structure of tRNA-Dependent Amidotransferase GatCAB | | Descriptor: | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit C, Glutamyl-tRNA(Gln) amidotransferase subunit A, ... | | Authors: | Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2006-02-23 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ammonia channel couples glutaminase with transamidase reactions in GatCAB
Science, 312, 2006
|
|
1DNM
 
 | | CRYSTAL STRUCTURE AND SEQUENCE-DEPENDENT CONFORMATION OF THE A.G MIS-PAIRED OLIGONUCLEOTIDE D(CGCAAGCTGGCG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*GP*CP*TP*GP*GP*CP*G)-3') | | Authors: | Webster, G.D, Sanderson, M.R, Skelly, J.V, Neidle, S, Swann, P.F, Li, B.F, Tickle, I.J. | | Deposit date: | 1990-06-22 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and sequence-dependent conformation of the A.G mispaired oligonucleotide d(CGCAAGCTGGCG).
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
1DC9
 
 | | PROPERTIES AND CRYSTAL STRUCTURE OF A BETA-BARREL FOLDING MUTANT, V60N INTESTINAL FATTY ACID BINDING PROTEIN (IFABP) | | Descriptor: | INTESTINAL FATTY ACID BINDING PROTEIN | | Authors: | Ropson, I.J, Yowler, B.C, Dalessio, P.M, Banaszak, L, Thompson, J. | | Deposit date: | 1999-11-04 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Properties and crystal structure of a beta-barrel folding mutant.
Biophys.J., 78, 2000
|
|
1DOV
 
 | |
