6MCS
 
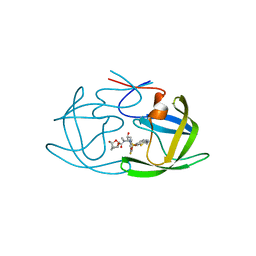 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-003 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(4-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, Protease | | Authors: | Bulut, H, Hayashi, H, Hattori, S.I, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2018-09-02 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Halogen Bond Interactions of Novel HIV-1 Protease Inhibitors (PI) (GRL-001-15 and GRL-003-15) with the Flap of Protease Are Critical for Their Potent Activity against Wild-Type HIV-1 and Multi-PI-Resistant Variants.
Antimicrob.Agents Chemother., 63, 2019
|
|
3BER
 
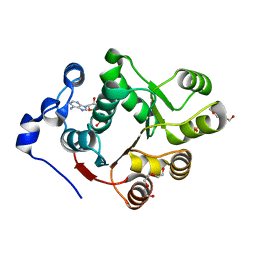 | | Human DEAD-box RNA-helicase DDX47, conserved domain I in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, PHOSPHATE ION, Probable ATP-dependent RNA helicase DDX47, ... | | Authors: | Karlberg, T, Busam, R.D, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-20 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
3BF6
 
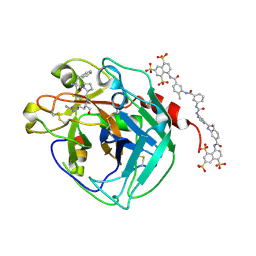 | | Thrombin:suramin complex | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFONIC ACID, PHE-PRO-ARG, Thrombin, ... | | Authors: | Lima, L.M.T.R, Polikarpov, I, Monteiro, R.Q. | | Deposit date: | 2007-11-20 | | Release date: | 2007-12-25 | | Last modified: | 2015-07-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and thermodynamic analysis of thrombin:suramin interaction in solution and crystal phases.
Biochim.Biophys.Acta, 1794, 2009
|
|
219L
 
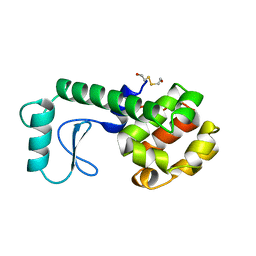 | | PROTEIN STRUCTURE PLASTICITY EXEMPLIFIED BY INSERTION AND DELETION MUTANTS IN T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, T4 LYSOZYME | | Authors: | Vetter, I.R, Baase, W.A, Heinz, D.W, Xiong, J.-P, Snow, S, Matthews, B.W. | | Deposit date: | 1996-09-23 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Protein structural plasticity exemplified by insertion and deletion mutants in T4 lysozyme.
Protein Sci., 5, 1996
|
|
4YZL
 
 | | Crystal structure of the indole prenyltransferase TleC complexed with indolactam V and DMSPP | | Descriptor: | (2S,5S)-5-(hydroxymethyl)-1-methyl-2-(propan-2-yl)-1,2,4,5,6,8-hexahydro-3H-[1,4]diazonino[7,6,5-cd]indol-3-one, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Tryptophan dimethylallyltransferase | | Authors: | Mori, T, Morita, H, Abe, I. | | Deposit date: | 2015-03-25 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Manipulation of prenylation reactions by structure-based engineering of bacterial indolactam prenyltransferases.
Nat Commun, 7, 2016
|
|
6MG4
 
 | | Structure of full-length human lambda-6A light chain JTO | | Descriptor: | JTO light chain | | Authors: | Morgan, G.J, Yan, N.L, Mortenson, D.E, Stanfield, R.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2018-09-12 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Stabilization of amyloidogenic immunoglobulin light chains by small molecules.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4YZT
 
 | | Crystal structure of a tri-modular GH5 (subfamily 4) endo-beta-1, 4-glucanase from Bacillus licheniformis complexed with cellotetraose | | Descriptor: | 1,2-ETHANEDIOL, Cellulose hydrolase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Liberato, M.V, Popov, A, Polikarpov, I. | | Deposit date: | 2015-03-25 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Molecular characterization of a family 5 glycoside hydrolase suggests an induced-fit enzymatic mechanism.
Sci Rep, 6, 2016
|
|
4Z2F
 
 | | Sclerotium Rolfsii lectin variant (SSR2) with fine carbohydrate specificity | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, lectin variant 2 | | Authors: | Kantsadi, A.L, Peppa, V.I, Leonidas, D.D. | | Deposit date: | 2015-03-29 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Cloning, Carbohydrate Specificity and the Crystal Structure of Two Sclerotium rolfsii Lectin Variants.
Molecules, 20, 2015
|
|
7JUE
 
 | |
3OUX
 
 | | Structure of beta-catenin with phosphorylated Lef-1 | | Descriptor: | Catenin beta-1, Lymphoid enhancer-binding factor 1 | | Authors: | Weis, W.I, Sun, J. | | Deposit date: | 2010-09-15 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and structural characterization of beta-catenin interactions with nonphosphorylated and CK2-phosphorylated Lef-1.
J.Mol.Biol., 405, 2011
|
|
6FSI
 
 | | Crystal structure of semiquinone Flavodoxin 1 from Bacillus cereus (1.32 A resolution) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, SULFATE ION, ... | | Authors: | Gudim, I, Lofstad, M, Hersleth, H.-P. | | Deposit date: | 2018-02-19 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | High-resolution crystal structures reveal a mixture of conformers of the Gly61-Asp62 peptide bond in an oxidized flavodoxin from Bacillus cereus.
Protein Sci., 27, 2018
|
|
3B7G
 
 | | Human DEAD-box RNA helicase DDX20, Conserved domain I (DEAD) in complex with AMPPNP (Adenosine-(Beta,gamma)-imidotriphosphate) | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable ATP-dependent RNA helicase DDX20 | | Authors: | Karlberg, T, Tresaugues, L, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Welin, M, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-30 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
4ZG0
 
 | | Crystal structure of Mouse Syndesmos protein | | Descriptor: | Protein syndesmos | | Authors: | Lee, I, Kim, H, Yoo, J, Cho, H, Lee, W. | | Deposit date: | 2015-04-22 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of syndesmos and its interaction with Syndecan-4 proteoglycan
Biochem.Biophys.Res.Commun., 463, 2015
|
|
4ZIQ
 
 | | Crystal structure of trypsin activated alpha-2-macroglobulin from Escherichia coli. | | Descriptor: | CHLORIDE ION, GLYCEROL, Uncharacterized lipoprotein YfhM | | Authors: | Garcia-Ferrer, I, Arede, P, Gomez-Blanco, J, Luque, D, Duquerroy, S, Caston, J.R, Goulas, T, Gomis-Ruth, X.F. | | Deposit date: | 2015-04-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and functional insights into Escherichia coli alpha 2-macroglobulin endopeptidase snap-trap inhibition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1NQC
 
 | | Crystal structures of Cathepsin S inhibitor complexes | | Descriptor: | Cathepsin S, N-[(1R)-2-(BENZYLSULFANYL)-1-FORMYLETHYL]-N-(MORPHOLIN-4-YLCARBONYL)-L-PHENYLALANINAMIDE | | Authors: | Pauly, T.A, Sulea, T, Ammirati, M, Sivaraman, J, Danley, D.E, Griffor, M.C, Kamath, A.V, Wang, I.K, Laird, E.R, Menard, R, Cygler, M, Rath, V.L. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Specificity determinants of human cathepsin s revealed
by crystal structures of complexes.
Biochemistry, 42, 2003
|
|
6C4Q
 
 | | 1.16 Angstrom Resolution Crystal Structure of Acyl Carrier Protein Domain (residues 1-100) of Polyketide Synthase Pks13 from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Polyketide synthase Pks13 | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-12 | | Release date: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | 1.16 Angstrom Resolution Crystal Structure of Acyl Carrier Protein Domain (residues 1-100) of Polyketide Synthase Pks13 from Mycobacterium tuberculosis.
To Be Published
|
|
4NYX
 
 | | Crystal Structure of the Bromodomain of human CREBBP in complex with a dihydroquinoxalinone ligand | | Descriptor: | (3R)-N-[3-(7-methoxy-3,4-dihydroquinolin-1(2H)-yl)propyl]-3-methyl-2-oxo-1,2,3,4-tetrahydroquinoxaline-5-carboxamide, CREB-binding protein | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Rooney, T.P.C, Fedorov, O, Martin, S, Monteiro, O.P, Conway, S.J, von Delft, F, Brennan, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-11 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structure of the Bromodomain of human CREBBP in complex with a dihydroquinoxalinone ligand
TO BE PUBLISHED
|
|
6FT1
 
 | | Crystal structure of oxidised Flavodoxin 1 from Bacillus cereus (1.4 A resolution) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, SULFATE ION, ... | | Authors: | Gudim, I, Lofstad, M, Hersleth, H.-P. | | Deposit date: | 2018-02-20 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution crystal structures reveal a mixture of conformers of the Gly61-Asp62 peptide bond in an oxidized flavodoxin from Bacillus cereus.
Protein Sci., 27, 2018
|
|
3P0L
 
 | | Human steroidogenic acute regulatory protein | | Descriptor: | Steroidogenic acute regulatory protein, mitochondrial | | Authors: | Lehtio, L, Siponen, M, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-29 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Comparative structural analysis of lipid binding START domains.
Plos One, 6, 2011
|
|
1NQS
 
 | | Structural Characterisation of the Holliday Junction formed by the sequence d(TCGGTACCGA) at 1.97 A | | Descriptor: | 5'-d(TpCpGpGpTpApCpCpGpA)-3', CALCIUM ION | | Authors: | Cardin, C.J, Gale, B.C, Thorpe, J.H, Texieira, S.C.M, Gan, Y, Moraes, M.I.A.A, Brogden, A.L. | | Deposit date: | 2003-01-22 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Analysis of two Holliday junctions formed by the sequences TCGGTACCGA and CCGGTACCGG
To be Published
|
|
6M3Y
 
 | | Crystal structure of pilus adhesin, SpaC from Lactobacillus rhamnosus GG - open conformation | | Descriptor: | MAGNESIUM ION, Pilus assembly protein | | Authors: | Kant, A, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2020-03-04 | | Release date: | 2020-07-29 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of lactobacillar SpaC reveals an atypical five-domain pilus tip adhesin: Exposing its substrate-binding and assembly in SpaCBA pili.
J.Struct.Biol., 211, 2020
|
|
6SNB
 
 | | Structure of Coxsackievirus A10 A-particle | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Zhao, Y, Zhou, D, Ni, T, Karia, D, Kotecha, A, Wang, X, Rao, Z, Jones, E.Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2019-08-23 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Hand-foot-and-mouth disease virus receptor KREMEN1 binds the canyon of Coxsackie Virus A10.
Nat Commun, 11, 2020
|
|
6MEE
 
 | | Crystal structure of broadly neutralizing antibody HEPC74 | | Descriptor: | antibody HEPC74 Heavy Chain, antibody HEPC74 Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J. | | Deposit date: | 2018-09-06 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | HCV Broadly Neutralizing Antibodies Use a CDRH3 Disulfide Motif to Recognize an E2 Glycoprotein Site that Can Be Targeted for Vaccine Design.
Cell Host Microbe, 24, 2018
|
|
4ZQA
 
 | |
6SP0
 
 | |
