5LGE
 
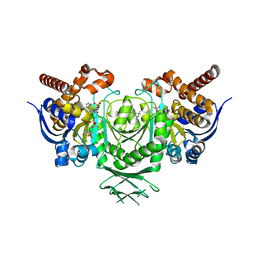 | | Crystal Structure of human IDH1 mutant (R132H) in complex with NADP+ and an Inhibitor related to BAY 1436032 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(4-propan-2-ylphenyl)amino]-1-[(1~{S},5~{S})-3,3,5-trimethylcyclohexyl]benzimidazole-5-carboxylic acid, ACETATE ION, ... | | Authors: | Hillig, R.C, Hars, U, Korndoerfer, I.P. | | Deposit date: | 2016-07-07 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pan-mutant IDH1 inhibitor BAY 1436032 for effective treatment of IDH1 mutant astrocytoma in vivo.
Acta Neuropathol., 133, 2017
|
|
4V6Y
 
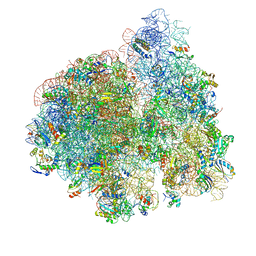 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in classic pre-translocation state (pre1a) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
5LT5
 
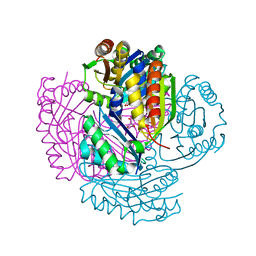 | | Carboxysome shell protein CcmP from Synechococcus elongatus PCC 7942 | | Descriptor: | CHLORIDE ION, CcmP, GLYCEROL | | Authors: | Larsson, A.M, Hasse, D, Valegard, K, Andersson, I. | | Deposit date: | 2016-09-06 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structures of beta-carboxysome shell protein CcmP: ligand binding correlates with the closed or open central pore.
J. Exp. Bot., 68, 2017
|
|
5EQG
 
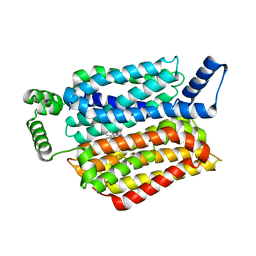 | | Human GLUT1 in complex with inhibitor (2~{S})-3-(4-fluorophenyl)-2-[2-(3-hydroxyphenyl)ethanoylamino]-~{N}-[(1~{S})-1-phenylethyl]propanamide | | Descriptor: | (2~{S})-3-(4-fluorophenyl)-2-[2-(3-hydroxyphenyl)ethanoylamino]-~{N}-[(1~{S})-1-phenylethyl]propanamide, Solute carrier family 2, facilitated glucose transporter member 1 | | Authors: | Kapoor, K, Finer-Moore, J, Pedersen, B.P, Caboni, L, Waight, A.B, Hillig, R, Bringmann, P, Heisler, I, Muller, T, Siebeneicher, H, Stroud, R.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of inhibition of human glucose transporter GLUT1 is conserved between cytochalasin B and phenylalanine amides.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2WF3
 
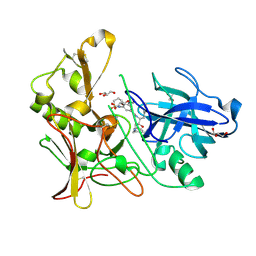 | | Human BACE-1 in complex with 6-(ethylamino)-N-((1S,2R)-2-hydroxy-3-(((3-(methyloxy)phenyl)methyl)amino)-1-(phenylmethyl)propyl)-1-methyl-1, 3,4,5-tetrahydro-2,1-benzothiazepine-8-carboxamide 2,2-dioxide | | Descriptor: | BETA-SECRETASE 1, GLYCEROL, N-{(1S,2R)-1-BENZYL-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}-6-(ETHYLAMINO)-1-METHYL-1,3,4,5-TETRAHYDRO-2,1-BENZOTHIAZEPINE-8-CARBOXAMIDE 2,2-DIOXIDE | | Authors: | Charrier, N, Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Hawkins, J, Hubbard, J, Hussain, I, Maile, G, Matico, R, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Rowland, P, Soleil, V, Smith, K.J, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Wayne, G. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-19 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Second Generation of Bace-1 Inhibitors Part 2: Optimisation of the Non-Prime Side Substituent.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2W1T
 
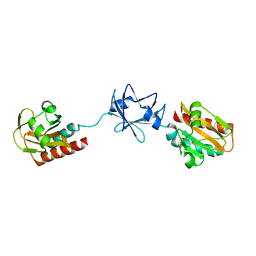 | | Crystal Structure of B. subtilis SpoVT | | Descriptor: | STAGE V SPORULATION PROTEIN T | | Authors: | Asen, I, Djuranovic, S, Lupas, A.N, Zeth, K. | | Deposit date: | 2008-10-20 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Spovt, the Final Modulator of Gene Expression During Spore Development in Bacillus Subtilis
J.Mol.Biol., 386, 2009
|
|
2VVY
 
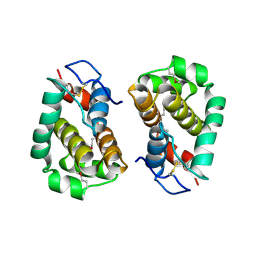 | | Structure of Vaccinia virus protein B14 | | Descriptor: | PROTEIN B15 | | Authors: | Graham, S.C, Bahar, M.W, Cooray, S, Chen, R.A.-J, Whalen, D.M, Abrescia, N.G.A, Alderton, D, Owens, R.J, Stuart, D.I, Smith, G.L, Grimes, J.M. | | Deposit date: | 2008-06-12 | | Release date: | 2008-08-26 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Vaccinia Virus Proteins A52 and B14 Share a Bcl-2-Like Fold But Have Evolved to Inhibit NF-kappaB Rather Than Apoptosis
Plos Pathog., 4, 2008
|
|
2VUX
 
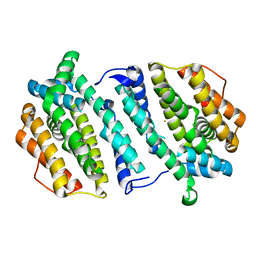 | | Human ribonucleotide reductase, subunit M2 B | | Descriptor: | FE (III) ION, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE SUBUNIT M2 B | | Authors: | Welin, M, Moche, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Svensson, L, Thorsell, A.G, Tresaugues, L, van Den Berg, S, Weigelt, J, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-31 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human Ribonucleotide Reductase, Subunit M2 B
To be Published
|
|
2WAH
 
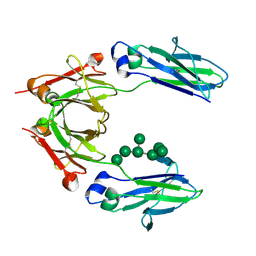 | | Crystal Structure of an IgG1 Fc Glycoform (Man9GlcNAc2) | | Descriptor: | IG GAMMA-1 CHAIN C REGION, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Crispin, M, Bowden, T.A, Coles, C.H, Harlos, K, Aricescu, A.R, Harvey, D.J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2009-02-06 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Carbohydrate and Domain Architecture of an Immature Antibody Glycoform Exhibiting Enhanced Effector Functions
J.Mol.Biol., 387, 2009
|
|
5LW6
 
 | |
2VSD
 
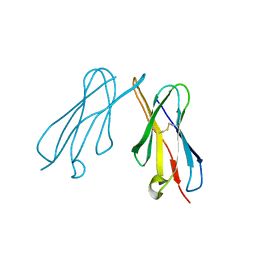 | | crystal structure of CHIR-AB1 | | Descriptor: | CHIR AB1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Arnon, T.I, Kaiser, J.T, Bjorkman, P.J. | | Deposit date: | 2008-04-22 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Crystal Structure of Chir-Ab1: A Primordial Avian Classical Fc Receptor.
J.Mol.Biol., 381, 2008
|
|
5LEG
 
 | | Structure of the bacterial sex F pilus (pED208) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, Pilin | | Authors: | Costa, T.R.D, Ilangovan, I, Ukleja, M, Redzej, A, Santini, J.M, Smith, T.K, Egelman, E.H, Waksman, G. | | Deposit date: | 2016-06-29 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the Bacterial Sex F Pilus Reveals an Assembly of a Stoichiometric Protein-Phospholipid Complex.
Cell, 166, 2016
|
|
5LCY
 
 | | Formaldehyde-Responsive Regulator FrmR E64H variant from Salmonella enterica serovar Typhimurium | | Descriptor: | Frmr | | Authors: | Pohl, E, Robinson, N, Osman, D, Piergentili, C, Uson, I. | | Deposit date: | 2016-06-22 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Effectors and Sensory Sites of Formaldehyde-responsive Regulator FrmR and Metal-sensing Variant.
J.Biol.Chem., 291, 2016
|
|
5EK8
 
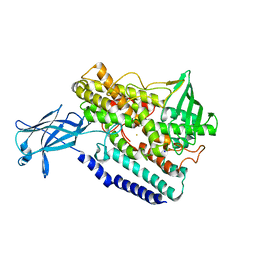 | | Crystal structure of a 9R-lipoxygenase from Cyanothece PCC8801 at 2.7 Angstroms | | Descriptor: | FE (II) ION, Lipoxygenase, SODIUM ION | | Authors: | Feussner, I, Ficner, R, Neumann, P, Newie, J, Andreou, A, Einsle, O. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a lipoxygenase from Cyanothece sp. may reveal novel features for substrate acquisition.
J.Lipid Res., 57, 2016
|
|
5EKJ
 
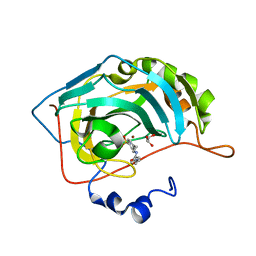 | |
4V77
 
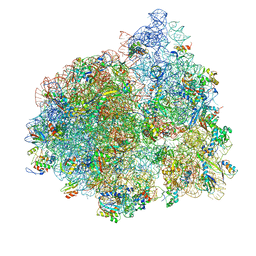 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in intermediate post-translocation state (post2b) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2W8X
 
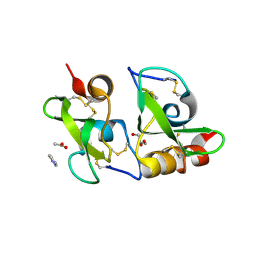 | | Structure of the tick ion-channel modulator Ra-KLP | | Descriptor: | ACETATE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Paesen, G.C, Siebold, C, Dallas, M, Peers, C, Harlos, K, Nuttall, P.A, Nunn, M.A, Stuart, D.I, Esnouf, R.M. | | Deposit date: | 2009-01-20 | | Release date: | 2009-05-05 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Ion-Channel Modulator from the Saliva of the Brown Ear Tick Has a Highly Modified Kunitz/Bpti Structure.
J.Mol.Biol., 389, 2009
|
|
4UXP
 
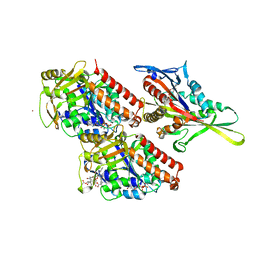 | | Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, KINESIN-3 MOTOR DOMAIN, ... | | Authors: | Atherton, J, Farabella, I, Yu, I.M, Rosenfeld, S.S, Houdusse, A, Topf, M, Moores, C. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Conserved Mechanisms of Microtubule-Stimulated Adp Release, ATP Binding, and Force Generation in Transport Kinesins.
Elife, 3, 2014
|
|
2W6Z
 
 | |
5EPE
 
 | | Crystal structure of SAM-dependent methyltransferase from Thiobacillus denitrificans in complex with S-Adenosyl-L-homocysteine | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase, SODIUM ION | | Authors: | LaRowe, C, Shabalin, I.G, Kutner, J, Handing, K.B, Stead, M, Hillerich, B.S, Ahmed, M, Seidel, R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-11-25 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of SAM-dependent methyltransferase from Thiobacillus denitrificans in complex with S-Adenosyl-L-homocysteine
to be published
|
|
2W05
 
 | | Structure of CDK2 in complex with an imidazolyl pyrimidine, compound 5b | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, N-(2-METHOXYETHYL)-4-({4-[2-METHYL-1-(1-METHYLETHYL)-1H-IMIDAZOL-5-YL]PYRIMIDIN-2-YL}AMINO)BENZENESULFONAMIDE | | Authors: | Anderson, M, Andrews, D.M, Barker, A.J, Brassington, C.A, Breed, J, Byth, K.F, Culshaw, J.D, Finlay, M.R, Fisher, E, Green, C.P, Heaton, D.W, Nash, I.A, Newcombe, N.J, Oakes, S.E, Pauptit, R.A, Roberts, A, Stanway, J.J, Thomas, A.P, Tucker, J.A, Weir, H.M. | | Deposit date: | 2008-08-08 | | Release date: | 2008-10-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Imidazoles: Sar and Development of a Potent Class of Cyclin-Dependent Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5M15
 
 | | Synthetic nanobody in complex with MBP | | Descriptor: | Maltose-binding periplasmic protein, Synthetic nanobody L2_D09, (a-MBP#3) | | Authors: | Zimmermann, I, Egloff, P, Seeger, M.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-11-15 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthetic single domain antibodies for the conformational trapping of membrane proteins.
Elife, 7, 2018
|
|
2W1R
 
 | | Crystal Structure of the C-terminal Domain of B. subtilis SpoVT | | Descriptor: | STAGE V SPORULATION PROTEIN T | | Authors: | Asen, I, Djuranovic, S, Lupas, A.N, Zeth, K. | | Deposit date: | 2008-10-20 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Spovt, the Final Modulator of Gene Expression During Spore Development in Bacillus Subtilis
J.Mol.Biol., 386, 2009
|
|
5LFB
 
 | | Structure of the bacterial sex F pilus (12.5 Angstrom rise) | | Descriptor: | Pilin, [(2~{S})-3-[[(2~{R})-2,3-bis(oxidanyl)propoxy]-oxidanyl-phosphoryl]oxy-2-hexadec-9-enoyloxy-propyl] hexadecanoate | | Authors: | Costa, T.R.D, Ilangovan, I, Ukleja, M, Redzej, A, Santini, J.M, Smith, T.K, Egelman, E.H, Waksman, G. | | Deposit date: | 2016-06-30 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structure of the Bacterial Sex F Pilus Reveals an Assembly of a Stoichiometric Protein-Phospholipid Complex.
Cell, 166, 2016
|
|
5EKM
 
 | |
