1YMN
 
 | | The study of reductive unfolding pathways of RNase A (Y92L mutant) | | Descriptor: | Ribonuclease pancreatic | | Authors: | Xu, G, Narayan, M, Kurinov, I, Ripoll, D.R, Welker, E, Khalili, M, Ealick, S.E, Scheraga, H.A. | | Deposit date: | 2005-01-21 | | Release date: | 2006-01-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A localized specific interaction alters the unfolding pathways of structural homologues.
J.Am.Chem.Soc., 128, 2006
|
|
7JW2
 
 | | Crystal structure of Aedes aegypti Nibbler EXO domain | | Descriptor: | Exonuclease mut-7 homolog | | Authors: | Xie, W, Sowemimo, I, Hayashi, R, Wang, J, Brennecke, J, Ameres, S.L, Patel, D.J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-function analysis of microRNA 3'-end trimming by Nibbler.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3L4B
 
 | | Crystal Structure of an Octomeric Two-Subunit TrkA K+ Channel Ring Gating Assembly, TM1088A:TM1088B, from Thermotoga maritima | | Descriptor: | ADENOSINE MONOPHOSPHATE, TrkA K+ Channel protein TM1088A, TrkA K+ Channel protein TM1088B, ... | | Authors: | Deller, M.C, Johnson, H.A, Miller, M, Spraggon, G, Wilson, I.A, Lesley, S.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2009-12-18 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal Structure of an Octomeric Two-Subunit TrkA K+ Channel Ring Gating Assembly, TM1088A:TM1088B, from Thermotoga maritima
To be Published
|
|
1I46
 
 | |
1I4C
 
 | |
1I8K
 
 | | CRYSTAL STRUCTURE OF DSFV MR1 IN COMPLEX WITH THE PEPTIDE ANTIGEN OF THE MUTANT EPIDERMAL GROWTH FACTOR RECEPTOR, EGFRVIII, AT LIQUID NITROGEN TEMPERATURE | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR ANTIBODY MR1SCFV HEAVY CHAIN, EPIDERMAL GROWTH FACTOR RECEPTOR ANTIBODY MR1SCFV LIGHT CHAIN, EPIDERMAL GROWTH FACTOR RECEPTOR, ... | | Authors: | Landry, R.C, Klimowicz, A.C, Lavictoire, S.J, Borisova, S, Kottachchi, D.T, Lorimer, I.A, Evans, S.V. | | Deposit date: | 2001-03-14 | | Release date: | 2002-03-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antibody recognition of a conformational epitope in a peptide antigen: Fv-peptide complex of an antibody fragment specific for the mutant EGF receptor, EGFRvIII.
J.Mol.Biol., 308, 2001
|
|
1IB7
 
 | | SOLUTION STRUCTURE OF F35Y MUTANT OF RAT FERRO CYTOCHROME B5, A CONFORMATION, ENSEMBLE OF 20 STRUCTURES | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shahzad, N, Dangi, B, Blankman, J.I, Guiles, R.D. | | Deposit date: | 2001-03-27 | | Release date: | 2001-04-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | MUTAGENIC MODULATION OF THE ENTROPY CHANGE ON OXIDATION OF CYTOCHROME B5: AN ANALYSIS OF THE CONTRIBUTION OF CONFORMATIONAL ENTROPY
To be Published
|
|
1X7N
 
 | | The crystal structure of Pyrococcus furiosus phosphoglucose isomerase with bound 5-phospho-D-arabinonate and Manganese | | Descriptor: | 5-PHOSPHOARABINONIC ACID, Glucose-6-phosphate isomerase, MANGANESE (II) ION | | Authors: | Berrisford, J.M, Akerboom, J, Brouns, S, Sedelnikova, S.E, Turnbull, A.P, van der Oost, J, Salmon, L, Hardre, R, Murray, I.A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2004-08-16 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The structures of inhibitor complexes of Pyrococcus furiosus phosphoglucose isomerase provide insights into substrate binding and catalysis.
J.Mol.Biol., 343, 2004
|
|
1ICR
 
 | | THE STRUCTURE OF ESCHERICHIA COLI NITROREDUCTASE COMPLEXED WITH NICOTINIC ACID | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINIC ACID, OXYGEN-INSENSITIVE NAD(P)H NITROREDUCTASE | | Authors: | Lovering, A.L, Hyde, E.I, Searle, P.F, White, S.A. | | Deposit date: | 2001-04-02 | | Release date: | 2001-04-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of Escherichia coli nitroreductase complexed with nicotinic acid: three crystal forms at 1.7 A, 1.8 A and 2.4 A resolution.
J.Mol.Biol., 309, 2001
|
|
1XAI
 
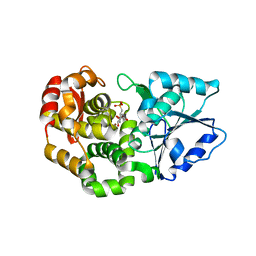 | | CRYSTAL STRUCTURE OF STAPHLYOCOCCUS AUREUS 3-DEHYDROQUINATE SYNTHASE (DHQS) IN COMPLEX WITH ZN2+, NAD+ AND CARBAPHOSPHONATE | | Descriptor: | 3-dehydroquinate synthase, ZINC ION, [1R-(1ALPHA,3BETA,4ALPHA,5BETA)]-5-(PHOSPHONOMETHYL)-1,3,4-TRIHYDROXYCYCLOHEXANE-1-CARBOXYLIC ACID | | Authors: | Nichols, C.E, Ren, J, Leslie, K, Dhaliwal, B, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-08-25 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Comparison of ligand induced conformational changes and domain closure mechanisms, between prokaryotic and eukaryotic dehydroquinate synthases.
J.Mol.Biol., 343, 2004
|
|
1XBC
 
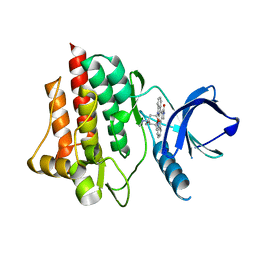 | | Crystal structure of the syk tyrosine kinase domain with Staurosporin | | Descriptor: | STAUROSPORINE, Tyrosine-protein kinase SYK | | Authors: | Badger, J, Atwell, S, Adams, J.M, Buchanan, M.D, Feil, I.K, Froning, K.J, Gao, X, Hendle, J, Keegan, K, Leon, B.C, Muller-Deickmann, H.J, Nienaber, V.L, Noland, B.W, Post, K, Rajashankar, K.R, Ramos, A, Russell, M, Burley, S.K, Buchanan, S.G. | | Deposit date: | 2004-08-30 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel mode of Gleevec binding is revealed by the structure of spleen tyrosine kinase
J.Biol.Chem., 279, 2004
|
|
1MR0
 
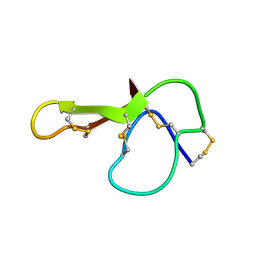 | | SOLUTION NMR STRUCTURE OF AGRP(87-120; C105A) | | Descriptor: | AGOUTI RELATED PROTEIN | | Authors: | Jackson, P.J, Mcnulty, J.C, Yang, Y.K, Thompson, D.A, Chai, B, Gantz, I, Barsh, G.S, Millhauser, G.M. | | Deposit date: | 2002-09-17 | | Release date: | 2002-10-02 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Design, pharmacology, and NMR structure of a minimized cystine knot with agouti-related protein activity.
Biochemistry, 41, 2002
|
|
1XRP
 
 | | Crystal structure of active site F1-mutant E213Q soaked with peptide Pro-Leu-Gly-Gly | | Descriptor: | PLGG, PROLINE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-15 | | Release date: | 2005-07-12 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
4FIA
 
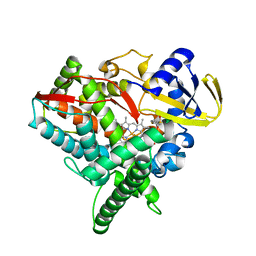 | | Crystal Structure of Human CYP46A1 P450 with bicalutamide Bound | | Descriptor: | Cholesterol 24-hydroxylase, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Stout, C.D, Pikuleva, I.A, Mast, N. | | Deposit date: | 2012-06-08 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of a cyano- and fluoro-containing drug bicalutamide to cytochrome P450 46A1: unusual features and spectral response.
J.Biol.Chem., 288, 2013
|
|
1XU4
 
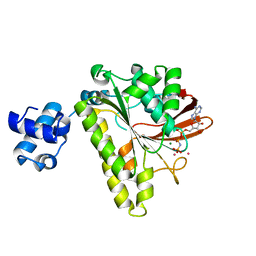 | | ATPASE IN COMPLEX WITH AMP-PNP, MAGNESIUM AND POTASSIUM CO-F | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Wu, Y, Qian, X, He, Y, Moya, I.A, Luo, Y. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an ATPase-active form of Rad51 homolog from Methanococcus voltae. Insights into potassium dependence
J.Biol.Chem., 280, 2005
|
|
3KSW
 
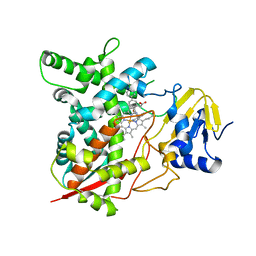 | | Crystal structure of sterol 14alpha-demethylase (CYP51) from Trypanosoma cruzi in complex with an inhibitor VNF ((4-(4-chlorophenyl)-N-[2-(1H-imidazol-1-yl)-1-phenylethyl]benzamide) | | Descriptor: | 4'-chloro-N-[(1R)-2-(1H-imidazol-1-yl)-1-phenylethyl]biphenyl-4-carboxamide, PROTOPORPHYRIN IX CONTAINING FE, Sterol 14-alpha demethylase | | Authors: | Lepesheva, G.I, Hargrove, T.Y, Anderson, S, Wawrzak, Z, Waterman, M.R. | | Deposit date: | 2009-11-23 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural Insights into Inhibition of Sterol 14{alpha}-Demethylase in the Human Pathogen Trypanosoma cruzi.
J.Biol.Chem., 285, 2010
|
|
1X8E
 
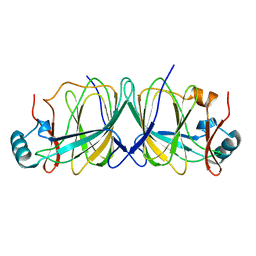 | | Crystal structure of Pyrococcus furiosus phosphoglucose isomerase free enzyme | | Descriptor: | Glucose-6-phosphate isomerase | | Authors: | Berrisford, J.M, Akerboom, J, Brouns, S, Sedelnikova, S.E, Turnbull, A.P, van der Oost, J, Salmon, L, Hardre, R, Murray, I.A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2004-08-18 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structures of inhibitor complexes of Pyrococcus furiosus phosphoglucose isomerase provide insights into substrate binding and catalysis.
J.Mol.Biol., 343, 2004
|
|
1MR8
 
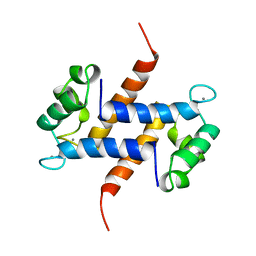 | | MIGRATION INHIBITORY FACTOR-RELATED PROTEIN 8 FROM HUMAN | | Descriptor: | CALCIUM ION, MIGRATION INHIBITORY FACTOR-RELATED PROTEIN 8 | | Authors: | Ishikawa, K, Nakagawa, A, Tanaka, I, Nishihira, J. | | Deposit date: | 1999-04-13 | | Release date: | 2000-05-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of human MRP8, a member of the S100 calcium-binding protein family, by MAD phasing at 1.9 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1I4B
 
 | |
1I5A
 
 | | STRUCTURE OF CHEA DOMAIN P4 IN COMPLEX WITH ADPCP AND MANGANESE | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Bilwes, A.M, Quezada, C.M, Croal, L.R, Crane, B.R, Simon, M.I. | | Deposit date: | 2001-02-26 | | Release date: | 2001-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleotide binding by the histidine kinase CheA.
Nat.Struct.Biol., 8, 2001
|
|
1N2Y
 
 | | SOLUTION STRUCTURE OF SS-CYCLIZED CATESTATIN FRAGMENT FROM CHROMOGRANIN A | | Descriptor: | CATESTATIN | | Authors: | Preece, N.E, Nguyen, M, Mahata, M, Mahata, S.K, Mahapatra, N.R, Tsigelny, I, O'Connor, D.T. | | Deposit date: | 2002-10-24 | | Release date: | 2002-11-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Conformational preferences and activities of peptides from the catecholamine
release-inhibitory (catestatin) region of chromogranin A
Regul.Pept., 118, 2004
|
|
1I8H
 
 | | SOLUTION STRUCTURE OF PIN1 WW DOMAIN COMPLEXED WITH HUMAN TAU PHOSPHOTHREONINE PEPTIDE | | Descriptor: | MICROTUBULE-ASSOCIATED PROTEIN TAU, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Wintjens, R, Wieruszeski, J.-M, Drobecq, H, Lippens, G, Landrieu, I. | | Deposit date: | 2001-03-14 | | Release date: | 2001-07-18 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | 1H NMR study on the binding of Pin1 Trp-Trp domain with phosphothreonine peptides.
J.Biol.Chem., 276, 2001
|
|
1XRE
 
 | | Crystal Structure of SodA-2 (BA5696) from Bacillus anthracis at 1.8A Resolution. | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Boucher, I.W, Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2004-10-14 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of two superoxide dismutases from Bacillus anthracis reveal a novel active centre.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1I31
 
 | | MU2 ADAPTIN SUBUNIT (AP50) OF AP2 CLATHRIN ADAPTOR, COMPLEXED WITH EGFR INTERNALIZATION PEPTIDE FYRALM AT 2.5 A RESOLUTION | | Descriptor: | CLATHRIN COAT ASSEMBLY PROTEIN AP50, EPIDERMAL GROWTH FACTOR RECEPTOR | | Authors: | Modis, Y, Boll, W, Rapoport, I, Kirchhausen, T. | | Deposit date: | 2001-02-12 | | Release date: | 2001-02-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MU2 ADAPTIN SUBUNIT (AP50) OF AP2 CLATHRIN ADAPTOR, COMPLEXED WITH EGFR INTERNALIZATION PEPTIDE FYRALM AT 2.5 A RESOLUTION
To be Published
|
|
3JQH
 
 | | Structure of the neck region of the glycan-binding receptor DC-SIGNR | | Descriptor: | C-type lectin domain family 4 member M | | Authors: | Feinberg, H, Tso, C.K.W, Taylor, M.E, Drickamer, K, Weis, W.I. | | Deposit date: | 2009-09-06 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Segmented helical structure of the neck region of the glycan-binding receptor DC-SIGNR.
J.Mol.Biol., 394, 2009
|
|
