7EYW
 
 | | Fe(II)/(alpha)ketoglutarate-dependent dioxygenase SptF with terretonin C | | Descriptor: | 2-oxoglutarate/Fe(II)-dependent dioxygenase SptF, FE (II) ION, N-OXALYLGLYCINE, ... | | Authors: | Tao, H, Mori, T, Abe, I. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular insights into the unusually promiscuous and catalytically versatile Fe(II)/ alpha-ketoglutarate-dependent oxygenase SptF.
Nat Commun, 13, 2022
|
|
7EYS
 
 | | Complex structure of SptF with Fe, alpha-ketoglutarate, and andiconin D | | Descriptor: | 2-OXOGLUTARIC ACID, 2-oxoglutarate/Fe(II)-dependent dioxygenase SptF, Andiconin D, ... | | Authors: | Tao, H, Mori, T, Abe, I. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular insights into the unusually promiscuous and catalytically versatile Fe(II)/ alpha-ketoglutarate-dependent oxygenase SptF.
Nat Commun, 13, 2022
|
|
8ABR
 
 | | Crystal structure of CYP109A2 from Bacillus megaterium bound with putative ligands hexanoic acid and octanoic acid | | Descriptor: | Cytochrome P450, HEME B/C, HEXANOIC ACID, ... | | Authors: | Jozwik, I.K, Rozeboom, H.J, Thunnissen, A.-M.W.H. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Regio- and stereoselective steroid hydroxylation by CYP109A2 from Bacillus megaterium explored by X-ray crystallography and computational modeling.
Febs J., 290, 2023
|
|
6YGU
 
 | | Crystal structure of the minimal Mtr4-Red1 complex (single chain) from Chaetomium thermophilum | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ATP dependent RNA helicase (Dob1)-like protein, ... | | Authors: | Dobrev, N, Ahmed, Y.L, Sinning, I. | | Deposit date: | 2020-03-27 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The zinc-finger protein Red1 orchestrates MTREC submodules and binds the Mtl1 helicase arch domain.
Nat Commun, 12, 2021
|
|
7EYT
 
 | | Fe(II)/(alpha)ketoglutarate-dependent dioxygenase SptF with andilesin C and NOG | | Descriptor: | 2-oxoglutarate/Fe(II)-dependent dioxygenase SptF, Andilesin C, FE (II) ION, ... | | Authors: | Tao, H, Mori, T, Abe, I. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular insights into the unusually promiscuous and catalytically versatile Fe(II)/ alpha-ketoglutarate-dependent oxygenase SptF.
Nat Commun, 13, 2022
|
|
6YRN
 
 | |
8A5S
 
 | | Crystal structure of light-activated DNA-binding protein EL222 from Erythrobacter litoralis crystallized in dark, measured illuminated. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Koval, T, Chaudhari, A, Fuertes, G, Andersson, I, Dohnalek, J. | | Deposit date: | 2022-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Light-dependent flavin redox and adduct states control the conformation and DNA-binding activity of the transcription factor EL222.
Nucleic Acids Res., 53, 2025
|
|
6YS4
 
 | |
6Y1I
 
 | |
6Y4A
 
 | | Crystal structure of the M295I variant of Ssl1 | | Descriptor: | COPPER (II) ION, Copper oxidase | | Authors: | Mielenbrink, S, Olbrich, A, Urlacher, V, Span, I. | | Deposit date: | 2020-02-20 | | Release date: | 2021-09-01 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.785 Å) | | Cite: | Substitution of the axial Type 1 Cu Ligand Afford Binding of a Water Molecule in Axial Position Affecting Kinetics, Spectral, and Structural Properties of the Small Laccase Ssl1.
Chemistry, 2024
|
|
6YAX
 
 | |
1KAN
 
 | |
9B33
 
 | |
9B35
 
 | | Ligand-binding and transmembrane domains of kainate receptor GluK2 in the open state, a complex with agonist glutamate and positive allosteric modulator BPAM344 | | Descriptor: | GLUTAMIC ACID, Glutamate receptor ionotropic, kainate 2 | | Authors: | Nadezhdin, K.D, Gangwar, S.P, Sobolevsky, A.I. | | Deposit date: | 2024-03-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Kainate receptor channel opening and gating mechanism.
Nature, 630, 2024
|
|
1KPA
 
 | | PKCI-1-ZINC | | Descriptor: | HUMAN PROTEIN KINASE C INTERACTING PROTEIN 1 (ZINC PROTEIN) | | Authors: | Lima, C.D, Klein, M.G, Weinstein, I.B, Hendrickson, W.A. | | Deposit date: | 1996-01-06 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of human protein kinase C interacting protein 1, a member of the HIT family of proteins.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1KPC
 
 | | PKCI-1-APO+ZINC | | Descriptor: | HUMAN PROTEIN KINASE C INTERACTING PROTEIN 1 (ZINC PROTEIN) | | Authors: | Lima, C.D, Klein, M.G, Weinstein, I.B, Hendrickson, W.A. | | Deposit date: | 1996-01-06 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of human protein kinase C interacting protein 1, a member of the HIT family of proteins.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1KPB
 
 | | PKCI-1-APO | | Descriptor: | HUMAN PROTEIN KINASE C INTERACTING PROTEIN 1 (ZINC PROTEIN) | | Authors: | Lima, C.D, Klein, M.G, Weinstein, I.B, Hendrickson, W.A. | | Deposit date: | 1996-01-06 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of human protein kinase C interacting protein 1, a member of the HIT family of proteins.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
6YU6
 
 | | Crystal structure of MhsT in complex with L-leucine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, LEUCINE, SODIUM ION, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
1KIV
 
 | |
6YU4
 
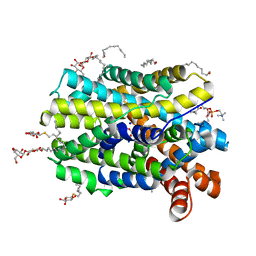 | | Crystal structure of MhsT in complex with L-4F-phenylalanine | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 4-FLUORO-L-PHENYLALANINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
1L2H
 
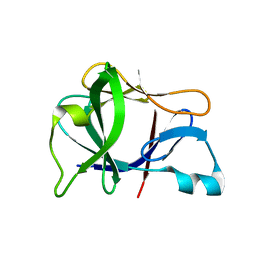 | | Crystal structure of Interleukin 1-beta F42W/W120F mutant | | Descriptor: | Interleukin 1-beta | | Authors: | Rudolph, M.G, Kelker, M.S, Schneider, T.R, Yeates, T.O, Oseroff, V, Heidary, D.K, Jennings, P.A, Wilson, I.A. | | Deposit date: | 2002-02-21 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Use of multiple anomalous dispersion to phase highly merohedrally twinned crystals of interleukin-1beta.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
8AIH
 
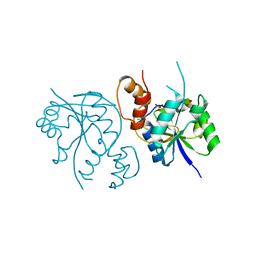 | | Crystal Structure of Enterococcus faecium Nicotinate Nucleotide Adenylyltransferase at 1.9 Angstroms Resolution | | Descriptor: | DIHYDROGENPHOSPHATE ION, Probable nicotinate-nucleotide adenylyltransferase, SULFATE ION | | Authors: | Pandian, R, Jeje, O.A, Sayed, Y, Achilonu, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Obtaining high yield recombinant Enterococcus faecium nicotinate nucleotide adenylyltransferase for X-ray crystallography and biophysical studies.
Int.J.Biol.Macromol., 250, 2023
|
|
1LGP
 
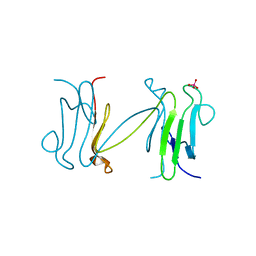 | | Crystal structure of the FHA domain of the Chfr mitotic checkpoint protein complexed with tungstate | | Descriptor: | TUNGSTATE(VI)ION, cell cycle checkpoint protein CHFR | | Authors: | Stavridi, E.S, Huyen, Y, Loreto, I.R, Scolnick, D.M, Halazonetis, T.D, Pavletich, N.P, Jeffrey, P.D. | | Deposit date: | 2002-04-16 | | Release date: | 2002-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the FHA domain of the Chfr mitotic checkpoint protein and its complex with tungstate.
Structure, 10, 2002
|
|
6Z72
 
 | | SARS-CoV-2 Macrodomain in complex with ADP-HPM | | Descriptor: | 1,2-ETHANEDIOL, Adenosine Diphosphate (Hydroxymethyl)pyrrolidine monoalcohol, D-MALATE, ... | | Authors: | Zorzini, V, Rack, J, Ahel, I. | | Deposit date: | 2020-05-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Viral macrodomains: a structural and evolutionary assessment of the pharmacological potential.
Open Biology, 10, 2020
|
|
6KYZ
 
 | | HRV14 3C in complex with single chain antibody YDF | | Descriptor: | Genome polyprotein, YDF H chain, YDF L chain | | Authors: | Meng, B, Yang, B, Wilson, I.A. | | Deposit date: | 2019-09-20 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Inhibitory antibodies identify unique sites of therapeutic vulnerability in rhinovirus and other enteroviruses.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
