8ZLQ
 
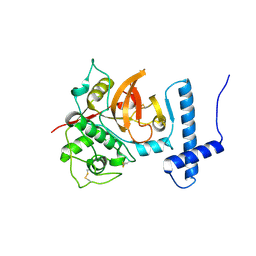 | | Proenzyme of Triticum aestivum papain-like cysteine protease Triticain-alpha inactive mutant lacking granulin domain | | Descriptor: | Triticain alpha | | Authors: | Petushkova, A.I, Zalevsky, A.O, Gorokhovets, N.V, Makarov, V.A, Maslova, V.D, Fedorov, V.A, Savvateeva, L.V, Zernii, E.Y, Soond, S.M, Golovin, A.V, Zamyatnin Jr, A.A. | | Deposit date: | 2024-05-21 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | The Structure of Triticain-Alpha Revealed a pH-Dependent Mechanism of Substrate Recognition
To Be Published
|
|
1KM8
 
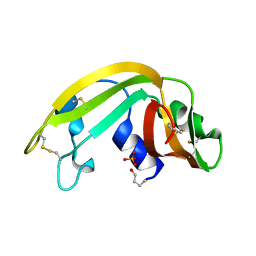 | | The Structure of a Cytotoxic Ribonuclease From the Oocyte of Rana Catesbeiana (Bullfrog) | | Descriptor: | PHOSPHATE ION, RIBONUCLEASE, OOCYTES | | Authors: | Chern, S.-S, Musayev, F.N, Amiraslanov, I.R, Liao, Y.-D, Liaw, Y.-C. | | Deposit date: | 2001-12-14 | | Release date: | 2003-09-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of a Cytotoxic Ribonuclease From the Oocyte of Rana Catesbeiana (Bullfrog)
To be Published
|
|
8RK1
 
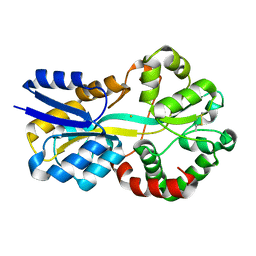 | | Crystal structure of FutA bound to Fe(III) solved by neutron diffraction | | Descriptor: | FE (III) ION, Putative iron ABC transporter, substrate binding protein | | Authors: | Bolton, R, Tews, I. | | Deposit date: | 2023-12-22 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-16 | | Method: | NEUTRON DIFFRACTION (2.095 Å) | | Cite: | A redox switch allows binding of Fe(II) and Fe(III) ions in the cyanobacterial iron-binding protein FutA from Prochlorococcus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8A53
 
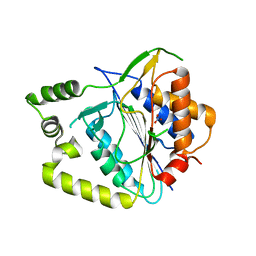 | | Crystal structure of AtMCA-IIf C147A (metacaspase 9) from Arabidopsis thaliana | | Descriptor: | Metacaspase-9, NITRATE ION | | Authors: | Sabljic, I, Stael, S, Stahlberg, J, Bozhkov, P. | | Deposit date: | 2022-06-14 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-function study of a Ca 2+ -independent metacaspase involved in lateral root emergence.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2VEN
 
 | | Structure-based enzyme engineering efforts with an inactive monomeric TIM variant: the importance of a single point mutation for generating an active site with suitable binding properties | | Descriptor: | CITRIC ACID, GLYCOSOMAL TRIOSEPHOSPHATE ISOMERASE | | Authors: | Alahuhta, M, Salin, M, Casteleijn, M.G, Kemmer, C, El-Sayed, I, Augustyns, K, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-10-25 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Protein Engineering Efforts with a Monomeric Tim Variant: The Importance of a Single Point Mutation for Generating an Active Site with Suitable Binding Properties.
Protein Eng.Des.Sel., 21, 2008
|
|
8ZNV
 
 | |
8ZNW
 
 | |
8S2V
 
 | |
8S2U
 
 | |
1KTX
 
 | | KALIOTOXIN (1-37) SHOWS STRUCTURAL DIFFERENCES WITH RELATED POTASSIUM CHANNEL BLOCKERS | | Descriptor: | KALIOTOXIN | | Authors: | Fernandez, I, Romi, R, Szendefi, S, Martin-Eauclaire, M.-F, Rochat, H, Van Rietschtoten, J, Pons, M, Giralt, E. | | Deposit date: | 1994-06-02 | | Release date: | 1995-01-26 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Kaliotoxin (1-37) shows structural differences with related potassium channel blockers.
Biochemistry, 33, 1994
|
|
8RXA
 
 | |
1KW1
 
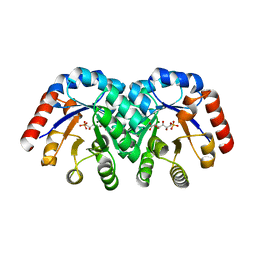 | | Crystal Structure of 3-Keto-L-Gulonate 6-Phosphate Decarboxylase with bound L-gulonate 6-phosphate | | Descriptor: | 3-Keto-L-Gulonate 6-Phosphate Decarboxylase, L-GULURONIC ACID 6-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E, Yew, W.S, Babbitt, P.C, Gerlt, J.A, Rayment, I. | | Deposit date: | 2002-01-28 | | Release date: | 2002-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Homologous (beta/alpha)8-barrel enzymes that catalyze unrelated reactions: orotidine 5'-monophosphate decarboxylase and 3-keto-L-gulonate 6-phosphate decarboxylase.
Biochemistry, 41, 2002
|
|
7FHS
 
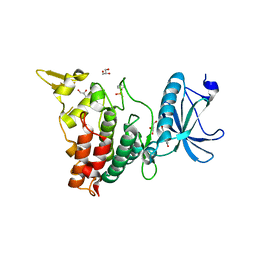 | | Crystal structure of DYRK1A in complex with RD0392 | | Descriptor: | (5~{Z})-5-[(3-ethoxy-4-oxidanyl-phenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, GLYCEROL | | Authors: | Kikuchi, M, Sumida, T, Hosoya, T, Kii, I, Umehara, T. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure-activity relationship for the folding intermediate-selective inhibition of DYRK1A.
Eur.J.Med.Chem., 227, 2022
|
|
7FHT
 
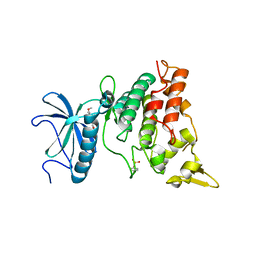 | | Crystal structure of DYRK1A in complex with RD0448 | | Descriptor: | (5~{Z})-5-[(3-ethynyl-4-methoxy-phenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Kikuchi, M, Sumida, Y, Hosoya, T, Kii, I, Umehara, T. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure-activity relationship for the folding intermediate-selective inhibition of DYRK1A.
Eur.J.Med.Chem., 227, 2022
|
|
1KXO
 
 | | ENGINEERED LIPOCALIN DIGA16 : APO-FORM | | Descriptor: | DigA16 | | Authors: | Korndoerfer, I.P, Skerra, A. | | Deposit date: | 2002-02-01 | | Release date: | 2003-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural mechanism of specific ligand recognition by a lipocalin tailored for the complexation of digoxigenin.
J.Mol.Biol., 330, 2003
|
|
8ZSW
 
 | | Crystal Structure of Human DDB1, a Component of the E3 Ubiquitin Ligase Complex | | Descriptor: | ACETATE ION, DNA damage-binding protein 1 | | Authors: | Lee, J, Gil, Y, Jeong, Y.R, Jo, I. | | Deposit date: | 2024-06-05 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Improved-Resolution Crystal Structure of Human DDB1, a Component of the E3 Ubiquitin Ligase Complex
To Be Published
|
|
7RSI
 
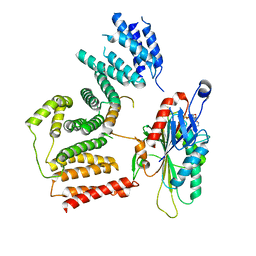 | | The cryo-EM map of KIF18A bound to KIFBP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KIF-binding protein, Kinesin-like protein KIF18A, ... | | Authors: | Tan, Z, Solon, A.L, Schutt, K.L, Jepsen, L, Haynes, S.E, Nesvizhskii, A.I, Sept, D, Stumpff, J, Ohi, R, Cianfrocco, M.A. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Kinesin-binding protein remodels the kinesin motor to prevent microtubule binding.
Sci Adv, 7, 2021
|
|
7RSQ
 
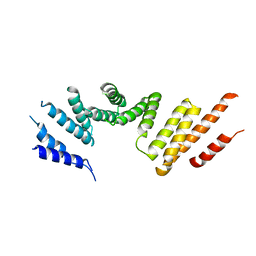 | | Cryo-EM structure of KIFBP core | | Descriptor: | KIF-binding protein | | Authors: | Solon, A.L, Tan, Z, Schutt, K.L, Jepsen, L, Haynes, S.E, Nesvizhskii, A.I, Sept, D, Stumpff, J, Ohi, R, Cianfrocco, M.A. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Kinesin-binding protein remodels the kinesin motor to prevent microtubule binding.
Sci Adv, 7, 2021
|
|
1KKH
 
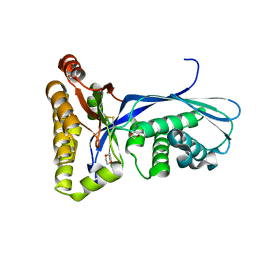 | | Crystal Structure of the Methanococcus jannaschii Mevalonate Kinase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Mevalonate Kinase | | Authors: | Yang, D, Shipman, L.W, Roessner, C.A, Scott, A.I, Sacchettini, J.C. | | Deposit date: | 2001-12-08 | | Release date: | 2002-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Methanococcus jannaschii mevalonate kinase, a member of the GHMP kinase superfamily.
J.Biol.Chem., 277, 2002
|
|
1KM9
 
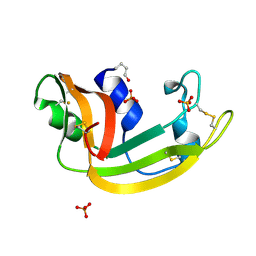 | | The Structure of a Cytotoxic Ribonuclease From the Oocyte of Rana Catesbeiana (Bullfrog) | | Descriptor: | PHOSPHATE ION, RIBONUCLEASE, OOCYTES | | Authors: | Chern, S.-S, Musayev, F.N, Amiraslanov, I.R, Liao, Y.-D, Liaw, Y.-C. | | Deposit date: | 2001-12-14 | | Release date: | 2003-09-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Structure of a Cytotoxic Ribonuclease From the Oocyte of Rana Catesbeiana (Bullfrog)
To be Published
|
|
8ABS
 
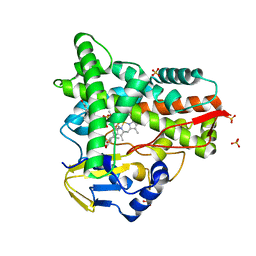 | | Crystal structure of CYP109A2 from Bacillus megaterium bound with testosterone and putative ligand 4,6-dimethyloctanoic acid | | Descriptor: | (4~{S},6~{S})-4,6-dimethyloctanoic acid, Cytochrome P450, HEME B/C, ... | | Authors: | Jozwik, I.K, Rozeboom, H.J, Thunnissen, A.-M.W.H. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Regio- and stereoselective steroid hydroxylation by CYP109A2 from Bacillus megaterium explored by X-ray crystallography and computational modeling.
Febs J., 290, 2023
|
|
6YO5
 
 | | Crystal structure of the M295F variant of Ssl1 | | Descriptor: | ALA-HIS-ALA, COPPER (II) ION, Copper oxidase, ... | | Authors: | Mielenbrink, S, Olbrich, A, Urlacher, V, Span, I. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substitution of the axial Type 1 Cu Ligand Afford Binding of a Water Molecule in Axial Position Affecting Kinetics, Spectral, and Structural Properties of the Small Laccase Ssl1.
Chemistry, 2024
|
|
7RYP
 
 | | Cryo-EM structure of KIFBP:KIF15 | | Descriptor: | KIF-binding protein, Kinesin-like protein KIF15 | | Authors: | Solon, A.L, Tan, Z, Schutt, K.L, Jepsen, L, Haynes, S.E, Nesvizhskii, A.I, Sept, D, Stumpff, J, Ohi, R, Cianfrocco, M.A. | | Deposit date: | 2021-08-25 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Kinesin-binding protein remodels the kinesin motor to prevent microtubule binding.
Sci Adv, 7, 2021
|
|
7EYR
 
 | | Fe(II)/(alpha)ketoglutarate-dependent dioxygenase SptF apo | | Descriptor: | 2-oxoglutarate/Fe(II)-dependent dioxygenase SptF, FE (II) ION | | Authors: | Tao, H, Mori, T, Abe, I. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Molecular insights into the unusually promiscuous and catalytically versatile Fe(II)/ alpha-ketoglutarate-dependent oxygenase SptF.
Nat Commun, 13, 2022
|
|
6KVF
 
 | | Structure of anti-hCXCR2 abN48 in complex with its CXCR2 epitope | | Descriptor: | Peptide from C-X-C chemokine receptor type 2, heavy chain, light chain | | Authors: | Xiang, J.C, Yan, L, Yang, B, Wilson, I.A. | | Deposit date: | 2019-09-04 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Selection of a picomolar antibody that targets CXCR2-mediated neutrophil activation and alleviates EAE symptoms.
Nat Commun, 12, 2021
|
|
