2IE0
 
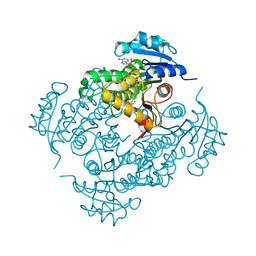 | | Crystal Structure of Isoniazid-resistant I21V Enoyl-ACP(COA) Reductase Mutant Enzyme From MYCOBACTERIUM TUBERCULOSIS in Complex with NADH-INH | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], ISONICOTINIC-ACETYL-NICOTINAMIDE-ADENINE DINUCLEOTIDE | | Authors: | Dias, M.V.B, Prado, A.M.X, Vasconceles, I.B, Fadel, F, Basso, L.A, Santos, D.S, Azevedo Jr, W.F. | | Deposit date: | 2006-09-15 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic studies on the binding of isonicotinyl-NAD adduct to wild-type and isoniazid resistant 2-trans-enoyl-ACP (CoA) reductase from Mycobacterium tuberculosis.
J.Struct.Biol., 159, 2007
|
|
5F44
 
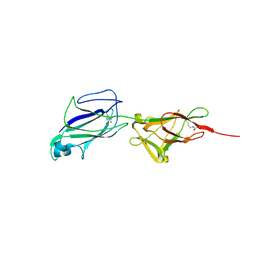 | | Crystal structure of shaft pilin spaA from Lactobacillus rhamnosus GG | | Descriptor: | ACETATE ION, Cell surface protein SpaA | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-03 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
7ZN5
 
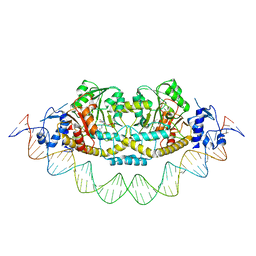 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C2 symmetry. | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-04-20 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
1F4I
 
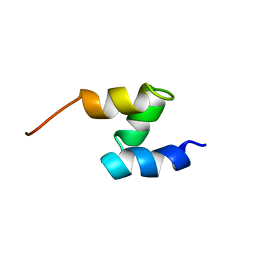 | | SOLUTION STRUCTURE OF THE HHR23A UBA(2) MUTANT P333E, DEFICIENT IN BINDING THE HIV-1 ACCESSORY PROTEIN VPR | | Descriptor: | UV EXCISION REPAIR PROTEIN PROTEIN RAD23 HOMOLOG A | | Authors: | Withers-Ward, E.S, Mueller, T.D, Chen, I.S, Feigon, J. | | Deposit date: | 2000-06-07 | | Release date: | 2000-12-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Biochemical and structural analysis of the interaction between the UBA(2) domain of the DNA repair protein HHR23A and HIV-1 Vpr.
Biochemistry, 39, 2000
|
|
7ZPA
 
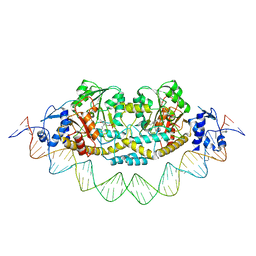 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C1 symmetry | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-04-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZLA
 
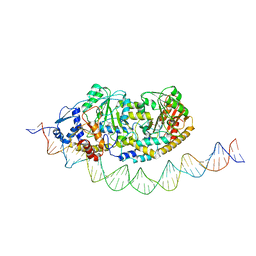 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the half-closed conformation | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Savino, C, Exertier, C, Bolognesi, M, Chaves Sanjuan, A. | | Deposit date: | 2022-04-14 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
1K3Y
 
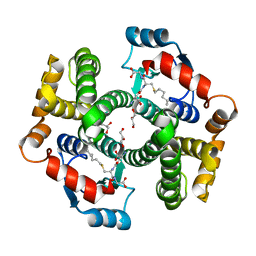 | | Crystal Structure Analysis of human Glutathione S-transferase with S-hexyl glutatione and glycerol at 1.3 Angstrom | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1, GLYCEROL, S-HEXYLGLUTATHIONE | | Authors: | Le Trong, I, Stenkamp, R.E, Ibarra, C, Atkins, W.M, Adman, E.T. | | Deposit date: | 2001-10-04 | | Release date: | 2002-10-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 1.3-A resolution structure of human glutathione S-transferase with S-hexyl glutathione bound reveals possible extended ligandin binding site.
Proteins, 48, 2002
|
|
1K4O
 
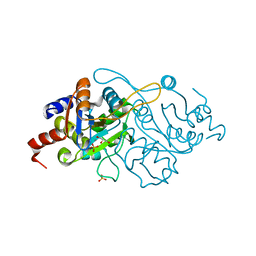 | | Crystal Structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase in complex with one Manganese, and a glycerol | | Descriptor: | 3,4-Dihydroxy-2-Butanone 4-Phosphate Synthase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Liao, D.-I, Zheng, Y.-J, Viitanen, P.V, Jordan, D.B. | | Deposit date: | 2001-10-08 | | Release date: | 2002-03-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural definition of the active site and catalytic mechanism of 3,4-dihydroxy-2-butanone-4-phosphate synthase.
Biochemistry, 41, 2002
|
|
3FN4
 
 | | Apo-form of NAD-dependent formate dehydrogenase from bacterium Moraxella sp.C-1 in closed conformation | | Descriptor: | GLYCEROL, NAD-dependent formate dehydrogenase, SULFATE ION | | Authors: | Shabalin, I.G, Polyakov, K.M, Filippova, E.V, Dorovatovskiy, P.V, Tikhonova, T.V, Sadykhov, E.G, Tishkov, V.I, Popov, V.O. | | Deposit date: | 2008-12-23 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of the apo and holo forms of formate dehydrogenase from the bacterium Moraxella sp. C-1: towards understanding the mechanism of the closure of the interdomain cleft
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1PSH
 
 | |
1U3R
 
 | | Crystal Structure of Estrogen Receptor beta complexed with WAY-338 | | Descriptor: | 2-(5-HYDROXY-NAPHTHALEN-1-YL)-1,3-BENZOOXAZOL-6-OL, Estrogen receptor beta, steroid receptor coactivator-1 | | Authors: | Malamas, M.S, Manas, E.S, McDevitt, R.E, Gunawan, I, Xu, Z.B, Collini, M.D, Miller, C.P, Dinh, T, Henderson, R.A, Keith Jr, J.C, Harris, H.A. | | Deposit date: | 2004-07-22 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Design and synthesis of aryl diphenolic azoles as potent and selective estrogen receptor-beta ligands.
J.Med.Chem., 47, 2004
|
|
6YFV
 
 | | Crystal structure of Mtr4-Red1 minimal complex from Chaetomium thermophilum | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ATP dependent RNA helicase (Dob1)-like protein, Red1, ... | | Authors: | Dobrev, N, Ahmed, Y.L, Sinning, I. | | Deposit date: | 2020-03-26 | | Release date: | 2021-05-05 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The zinc-finger protein Red1 orchestrates MTREC submodules and binds the Mtl1 helicase arch domain.
Nat Commun, 12, 2021
|
|
6YRL
 
 | |
5EZP
 
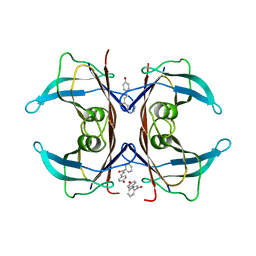 | | Human transthyretin (TTR) complexed with 4-hydroxy-chalcone | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-chalcone, Transthyretin | | Authors: | Polsinelli, I, Nencetti, S, Shepard, W.E, Orlandini, E, Stura, E.A. | | Deposit date: | 2015-11-26 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new crystal form of human transthyretin obtained with a curcumin derived ligand.
J.Struct.Biol., 194, 2016
|
|
5T1Q
 
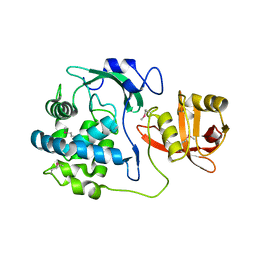 | | 2.15 Angstrom Crystal Structure of N-acetylmuramoyl-L-alanine Amidase from Staphylococcus aureus. | | Descriptor: | N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979, SODIUM ION, TRIETHYLENE GLYCOL | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bagnoli, F, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-19 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.15 Angstrom Crystal Structure of N-acetylmuramoyl-L-alanine Amidase from Staphylococcus aureus.
To Be Published
|
|
5T28
 
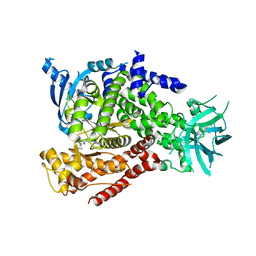 | | mPI3Kd IN COMPLEX WITH 5k | | Descriptor: | 3-(benzotriazol-1-yl)-5-[1-methyl-5-(1-phenylcyclopropyl)-1,2,4-triazol-3-yl]pyrazin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Petersen, J, Terstige, I, Perry, M, Svensson, T, Tyrchan, C, Lindmark, H, Oster, L. | | Deposit date: | 2016-08-23 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of triazole aminopyrazines as a highly potent and selective series of PI3K delta inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
1K49
 
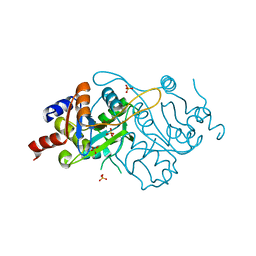 | | Crystal Structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase (cation free form) | | Descriptor: | 3,4-Dihydroxy-2-Butanone 4-Phosphate Synthase, SULFATE ION | | Authors: | Liao, D.-I, Zheng, Y.-J, Viitanen, P.V, Jordan, D.B. | | Deposit date: | 2001-10-06 | | Release date: | 2002-03-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural definition of the active site and catalytic mechanism of 3,4-dihydroxy-2-butanone-4-phosphate synthase.
Biochemistry, 41, 2002
|
|
2IN6
 
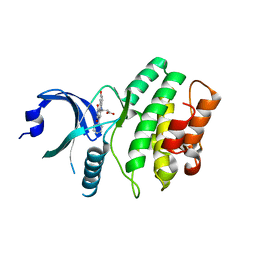 | | Wee1 kinase complex with inhibitor PD311839 | | Descriptor: | 3-(9-HYDROXY-1,3-DIOXO-4-PHENYL-2,3-DIHYDROPYRROLO[3,4-C]CARBAZOL-6(1H)-YL)PROPANOIC ACID, Wee1-like protein kinase | | Authors: | Squire, C.J, Dickson, J.M, Ivanovic, I, Baker, E.N. | | Deposit date: | 2006-10-05 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and structure-activity relationships of N-6 substituted analogues of 9-hydroxy-4-phenylpyrrolo[3,4-c]carbazole-1,3(2H,6H)-diones as inhibitors of Wee1 and Chk1 checkpoint kinases.
Eur.J.Med.Chem., 43, 2008
|
|
1AFD
 
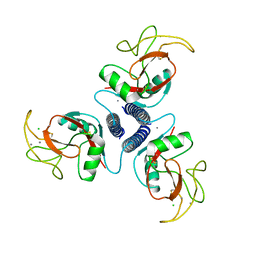 | |
4D2G
 
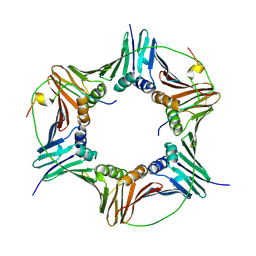 | | Crystal structure of human PCNA in complex with p15 peptide | | Descriptor: | P15, PROLIFERATING CELL NUCLEAR ANTIGEN | | Authors: | DeBiasio, A, Ibanez, A, Mortuza, G, Molina, R, Cordeiro, T.N, Castillo, F, Villate, M, Merino, N, Lelli, M, Diercks, T, Luque, I, Bernardo, P, Montoya, G, Blanco, F.J. | | Deposit date: | 2014-05-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of P15(Paf)-PCNA Complex and Implications for Clamp Sliding During DNA Replication and Repair.
Nat.Commun., 6, 2015
|
|
6E2J
 
 | | Crystal structure of the heterocomplex between human keratin 1 coil 1B containing S233L mutation and wild-type human keratin 10 coil 1B | | Descriptor: | Keratin, type I cytoskeletal 10, type II cytoskeletal 1, ... | | Authors: | Eldirany, S.A, Lomakin, I.B, Bunick, C.G. | | Deposit date: | 2018-07-11 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.386 Å) | | Cite: | Human keratin 1/10-1B tetramer structures reveal a knob-pocket mechanism in intermediate filament assembly.
Embo J., 38, 2019
|
|
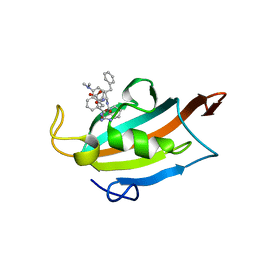
6RCY
 
 | |
2V23
 
 | | Structure of cytochrome c peroxidase mutant N184R Y36A | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Metcalfe, C.L, Macdonald, I.K, Brown, K.A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2007-05-31 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Tuberculosis Prodrug Isoniazid Bound to Activating Peroxidases.
J.Biol.Chem., 283, 2008
|
|
7ZM5
 
 | | Structure of Mossman virus receptor binding protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein | | Authors: | Stelfox, A.J, Bowden, T.A, Rissanen, I, Harlos, K. | | Deposit date: | 2022-04-19 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure and solution state of the C-terminal head region of the narmovirus receptor binding protein.
Mbio, 14, 2023
|
|
7ZM6
 
 | | Nariva virus receptor binding protein | | Descriptor: | Attachment protein | | Authors: | Stelfox, A.J, Rissanen, I, Rambo, R, Lee, B, Bowden, T.A. | | Deposit date: | 2022-04-19 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure and solution state of the C-terminal head region of the narmovirus receptor binding protein.
Mbio, 14, 2023
|
|
