2FTK
 
 | | berylloflouride Spo0F complex with Spo0B | | Descriptor: | MAGNESIUM ION, Sporulation initiation phosphotransferase B, Sporulation initiation phosphotransferase F | | Authors: | Varughese, K.I. | | Deposit date: | 2006-01-24 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The Crystal Structure of Beryllofluoride Spo0F in Complex with the Phosphotransferase Spo0B Represents a Phosphotransfer Pretransition State.
J.Bacteriol., 188, 2006
|
|
6HCS
 
 | |
6H9J
 
 | | Factor Inhibiting HIF (FIH) in complex with zinc, NOG and TRPV3 (229-255) | | Descriptor: | GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, N-OXALYLGLYCINE, ... | | Authors: | Leissing, T.M, Clifton, I.J, Saward, B.G, Lu, X, Hopkinson, R.J, Schofield, C.J. | | Deposit date: | 2018-08-04 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Factor Inhibiting HIF (FIH) in complex with zinc, NOG and TRPV3 (229-255)
To Be Published
|
|
3HW7
 
 | | High pressure (0.57 GPa) crystal structure of bovine copper, zinc superoxide dismutase at 2.0 angstroms | | Descriptor: | COPPER (I) ION, COPPER (II) ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Ascone, I, Savino, C. | | Deposit date: | 2009-06-17 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flexibility of the Cu,Zn superoxide dismutase structure investigated at 0.57 GPa
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3PGM
 
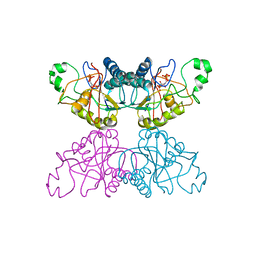 | | THE STRUCTURE OF YEAST PHOSPHOGLYCERATE MUTASE AT 0.28 NM RESOLUTION | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Phosphoglycerate mutase 1, SULFATE ION | | Authors: | Campbell, J.W, Hodgson, G.I, Warwicker, J, Winn, S.I, Watson, H.C. | | Deposit date: | 1982-04-06 | | Release date: | 1982-05-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and activity of phosphoglycerate mutase.
Philos.Trans.R.Soc.London,Ser.B, 293, 1981
|
|
4NM4
 
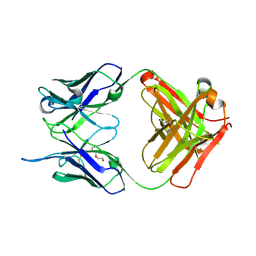 | |
6V4C
 
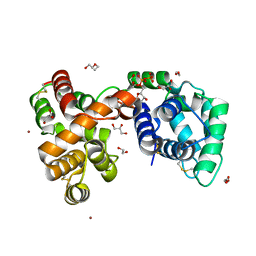 | | Culex quinquefasciatus D7 long form 1- CxD7L1 in complex with ADP | | Descriptor: | 1,2-ETHANEDIOL, 2-ETHOXYETHANOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Calvo, E, Garboczi, D.N, Martin-Martin, I, Gittis, A.G. | | Deposit date: | 2019-11-27 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | ADP binding by the Culex quinquefasciatus mosquito D7 salivary protein enhances blood feeding on mammals.
Nat Commun, 11, 2020
|
|
6FVT
 
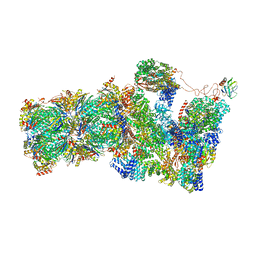 | | 26S proteasome, s1 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
6FXE
 
 | | BBE31 from Lyme disease agent Borrelia (Borreliella) burgdorferi playing a vital role in successful colonization of the mammalian host | | Descriptor: | GLUTATHIONE, Putative surface protein | | Authors: | Brangulis, K, Akopjana, I, Petrovskis, I, Kazaks, A, Tars, K. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-20 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | BBE31 from the Lyme disease agent Borrelia burgdorferi, known to play an important role in successful colonization of the mammalian host, shows the ability to bind glutathione.
Biochim Biophys Acta Gen Subj, 1864, 2019
|
|
6MLY
 
 | | Bifunctional GH43-CE Bacteroides eggerthii, BACEGG_01304 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Bifunctional GH43-CE protein, ... | | Authors: | Koropatkin, N.M, Pereira, G.V, Cann, I. | | Deposit date: | 2018-09-28 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Degradation of complex arabinoxylans by human colonic Bacteroidetes
Nat Commun, 2021
|
|
6RTP
 
 | |
8C1V
 
 | | SARS-CoV-2 S-trimer (3 RBDs up) bound to TriSb92, fitted into cryo-EM map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sb92, ... | | Authors: | Huiskonen, J.T, Rissanen, I, Hannula, L. | | Deposit date: | 2022-12-21 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Intranasal trimeric sherpabody inhibits SARS-CoV-2 including recent immunoevasive Omicron subvariants.
Nat Commun, 14, 2023
|
|
6N4X
 
 | | Metabotropic Glutamate Receptor 5 Apo Form Ligand Binding Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Metabotropic glutamate receptor 5 | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Weis, W.I, Skiniotis, G.S, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
2H8P
 
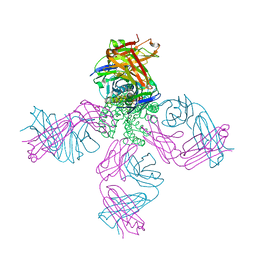 | | Structure of a K channel with an amide to ester substitution in the selectivity filter | | Descriptor: | (2S)-2-(BUTYRYLOXY)-3-HYDROXYPROPYL NONANOATE, FAB heavy chain, FAB light chain, ... | | Authors: | Valiyaveetil, F.I, MacKinnon, R, Muir, T.W. | | Deposit date: | 2006-06-07 | | Release date: | 2006-09-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and Functional Consequences of an Amide-to-Ester Substitution in the Selectivity Filter of a Potassium Channel.
J.Am.Chem.Soc., 128, 2006
|
|
2O6H
 
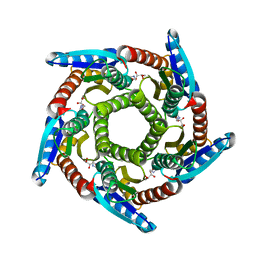 | | Lumazine synthase RibH1 from Brucella melitensis (Gene BMEI1187, Swiss-Prot entry Q8YGH2) complexed with inhibitor 5-Nitro-6-(D-Ribitylamino)-2,4(1H,3H) Pyrimidinedione | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase 1, CALCIUM ION | | Authors: | Klinke, S, Zylberman, V, Bonomi, H.R, Haase, I, Guimaraes, B.G, Braden, B.C, Bacher, A, Fischer, M, Goldbaum, F.A. | | Deposit date: | 2006-12-07 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Kinetic Properties of Lumazine Synthase Isoenzymes in the Order Rhizobiales
J.Mol.Biol., 373, 2007
|
|
1JPP
 
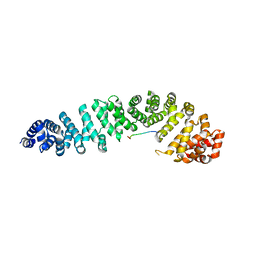 | |
6S7U
 
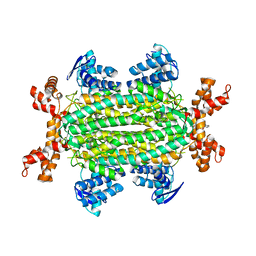 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(5-(Azepan-1-ylsulfonyl)-2-methoxyphenyl)-2-(1H-indol-3-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, MAGNESIUM ION, ... | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-07-06 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
2AOC
 
 | | Crystal structure analysis of HIV-1 protease mutant I84V with a substrate analog P2-NC | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
3N9X
 
 | | Crystal structure of Map Kinase from plasmodium berghei, PB000659.00.0 | | Descriptor: | GLYCEROL, Phosphotransferase | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, MacKenzie, F, Kozieradzki, I, Chau, I, Lew, J, Senisterra, G, Artz, J, Amani, M, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-31 | | Release date: | 2010-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Map Kinase from plasmodium berghei, PB000659.00.0
To be Published
|
|
4P3C
 
 | | MT1-MMP:Fab complex (Form I) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Rozenberg, H, Udi, Y, Sagi, I. | | Deposit date: | 2014-03-06 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Inhibition mechanism of membrane metalloprotease by an exosite-swiveling conformational antibody.
Structure, 23, 2015
|
|
5K5T
 
 | | Crystal structure of the inactive form of human calcium-sensing receptor extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular calcium-sensing receptor, ... | | Authors: | Geng, Y, Mosyak, L, Kurinov, I, Zuo, H, Sturchler, E, Cheng, T.C, Subramanyam, P, Brown, A.P, Brennan, S.C, Mun, H.-C, Bush, M, Chen, Y, Nguyen, T, Cao, B, Chang, D, Quick, M, Conigrave, A, Colecraft, H.M, McDonald, P, Fan, Q.R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural mechanism of ligand activation in human calcium-sensing receptor.
Elife, 5, 2016
|
|
6N32
 
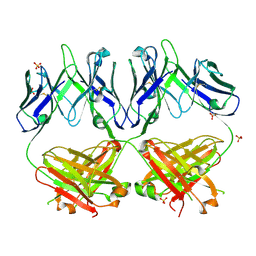 | | Anti-HIV-1 Fab 2G12 re-refinement | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, SULFATE ION | | Authors: | Calarese, D.A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-11-14 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antibody domain exchange is an immunological solution to carbohydrate cluster recognition.
Science, 300, 2003
|
|
2VDC
 
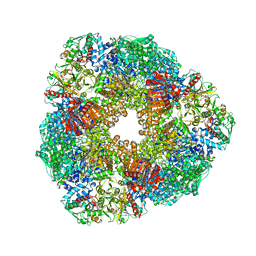 | | THE 9.5 A RESOLUTION STRUCTURE OF GLUTAMATE SYNTHASE FROM CRYO-ELECTRON MICROSCOPY AND ITS OLIGOMERIZATION BEHAVIOR IN SOLUTION: FUNCTIONAL IMPLICATIONS. | | Descriptor: | 2-OXOGLUTARIC ACID, FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Cottevieille, M, Larquet, E, Jonic, S, Petoukhov, M.V, Caprini, G, Paravisi, S, Svergun, D.I, Vanoni, M.A, Boisset, N. | | Deposit date: | 2007-10-04 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | The Subnanometer Resolution Structure of the Glutamate Synthase 1.2-Mda Hexamer by Cryoelectron Microscopy and its Oligomerization Behavior in Solution: Functional Implications.
J.Biol.Chem., 283, 2008
|
|
3L8Y
 
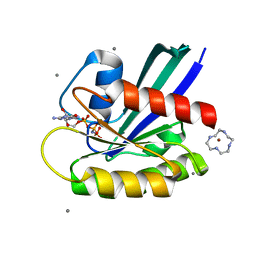 | | Complex of Ras with cyclen | | Descriptor: | 1,4,7,10-tetraazacyclododecane, CALCIUM ION, GTPase HRas, ... | | Authors: | Rosnizeck, I.C, Graf, T, Spoerner, M, Traenkle, J, Filchtinski, D, Herrmann, C, Gremer, L, Vetter, I.R, Wittinghofer, A, Koenig, B, Kalbitzer, H.R. | | Deposit date: | 2010-01-04 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Stabilizing a weak binding state for effectors in the human ras protein by cyclen complexes
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
6N51
 
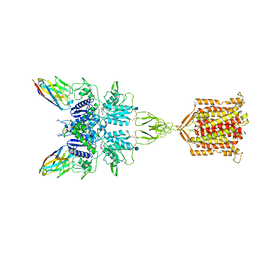 | | Metabotropic Glutamate Receptor 5 bound to L-quisqualate and Nb43 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5, ... | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Weis, W.I, Skiniotis, G.S, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
