6E9A
 
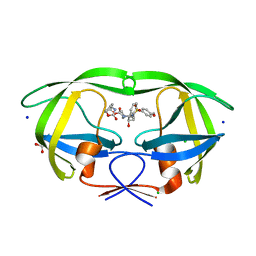 | | HIV-1 WILD TYPE PROTEASE WITH GRL-034-17A, (3aS, 5R, 6aR)-2-OXOHEXAHYD CYCLOPENTA[D]-5-OXAZOLYL URETHANE WITH A BICYCLIC OXAZOLIDINONE SCAFF AS THE P2 LIGAND | | Descriptor: | (3aS,5R,6aR)-2-oxohexahydro-2H-cyclopenta[d][1,3]oxazol-5-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2018-07-31 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Design and Synthesis of Potent HIV-1 Protease Inhibitors Containing Bicyclic Oxazolidinone Scaffold as the P2 Ligands: Structure-Activity Studies and Biological and X-ray Structural Studies.
J. Med. Chem., 61, 2018
|
|
7RZA
 
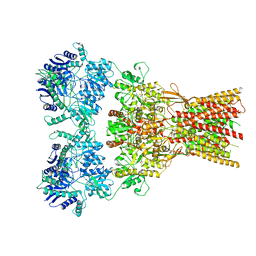 | | Structure of the complex of AMPA receptor GluA2 with auxiliary subunit GSG1L bound to agonist quisqualate | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2 | | Authors: | Gangwar, S.P, Klykov, O.V, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-27 | | Release date: | 2021-10-27 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
7RYZ
 
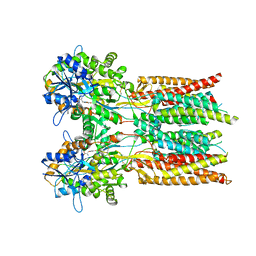 | | Structure of the complex of LBD-TMD part of AMPA receptor GluA2 with auxiliary subunit GSG1L bound to agonist quisqualate | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2 | | Authors: | Gangwar, S.P, Klykov, O.V, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-26 | | Release date: | 2021-10-27 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
7S4Y
 
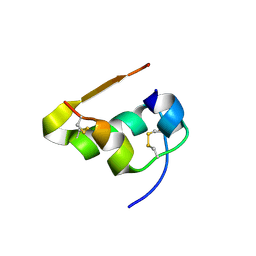 | | Serial Macromolecular Crystallography at ALBA Synchrotron Light Source - Insulin | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Martin-Garcia, J.M, Botha, S, Hu, H, Jernigan, R, Castellvi, A, Lisova, S, Gil, F, Calisto, B, Crespo, I, Roy-Chowdbury, S, Grieco, A, Ketawala, G, Weierstall, U, Spence, J, Fromme, P, Zatsepin, N, Boer, R, Carpena, X. | | Deposit date: | 2021-09-09 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Serial macromolecular crystallography at ALBA Synchrotron Light Source.
J.Synchrotron Radiat., 29, 2022
|
|
5AA3
 
 | |
1JYM
 
 | | Crystals of Peptide Deformylase from Plasmodium falciparum with Ten Subunits per Asymmetric Unit Reveal Critical Characteristics of the Active Site for Drug Design | | Descriptor: | COBALT (II) ION, Peptide Deformylase | | Authors: | Kumar, A, Nguyen, K.T, Srivathsan, S, Ornstein, B, Turley, S, Hirsh, I, Pei, D, Hol, W.G.J. | | Deposit date: | 2001-09-12 | | Release date: | 2002-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystals of peptide deformylase from Plasmodium falciparum reveal critical characteristics of the active site for drug design.
Structure, 10, 2002
|
|
7S50
 
 | | Serial Macromolecular Crystallography at ALBA Synchrotron Light Source - Phycocyanin | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, CHLORIDE ION, ... | | Authors: | Martin-Garcia, J.M, Botha, S, Hu, H, Jernigan, R, Castellvi, A, Lisova, S, Gil, F, Calisto, B, Crespo, I, Roy-Chowdbury, S, Grieco, A, Ketawala, G, Weierstall, U, Spence, J, Fromme, P, Zatsepin, N, Boer, R, Carpena, X. | | Deposit date: | 2021-09-09 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial macromolecular crystallography at ALBA Synchrotron Light Source.
J.Synchrotron Radiat., 29, 2022
|
|
7S6G
 
 | | Crystal structure of PhnD from Synechococcus MITS9220 in complex with phosphate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Shah, B.S, Mikolajek, H, Orr, C.M, Mykhaylyk, V, Owens, R.J, Paulsen, I.T. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Marine picocyanobacterial PhnD1 shows specificity for various phosphorus sources but likely represents a constitutive inorganic phosphate transporter.
Isme J, 17, 2023
|
|
7RZ9
 
 | |
6ZB5
 
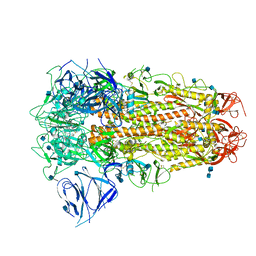 | | SARS CoV-2 Spike protein, Closed conformation, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Burucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2020-06-07 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein.
Science, 370, 2020
|
|
6Z8O
 
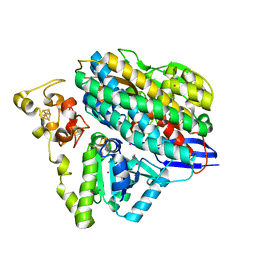 | | Structure of [NiFeSe] hydrogenase G491A variant from Desulfovibrio vulgaris Hildenborough pressurized with Krypton gas - structure G491A-Kr | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE (II) ION, ... | | Authors: | Zacarias, S, Temporao, A, Carpentier, P, van der Linden, P, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2020-06-02 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring the gas access routes in a [NiFeSe] hydrogenase using crystals pressurized with krypton and oxygen.
J.Biol.Inorg.Chem., 25, 2020
|
|
7RZ7
 
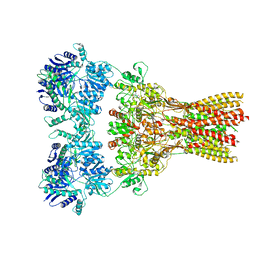 | | Structure of the complex of AMPA receptor GluA2 with auxiliary subunit TARP gamma-5 bound to agonist Quisqualate | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2 | | Authors: | Klykov, O.V, Gangwar, S.P, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-27 | | Release date: | 2021-10-27 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
6ZB4
 
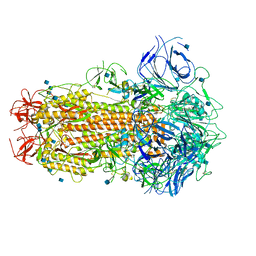 | | SARS CoV-2 Spike protein, Closed conformation, C1 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Burucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein.
Science, 370, 2020
|
|
6YU7
 
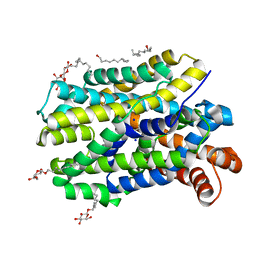 | | Crystal structure of MhsT in complex with L-tyrosine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, SODIUM ION, Sodium-dependent transporter, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
2W35
 
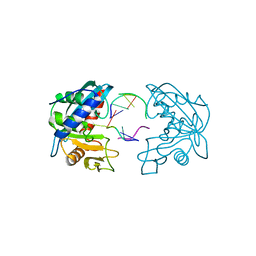 | | Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair | | Descriptor: | 5'-D(*AP*GP*CP*CP*GP*TP)-3', 5'-D(*AP*TP*GP*CP*GP*AP*CP*IP*GP)-3', Endonuclease V, ... | | Authors: | Dalhus, B, Arvai, A.S, Rosnes, I, Olsen, O.E, Backe, P.H, Alseth, I, Gao, H, Cao, W, Tainer, J.A, Bjoras, M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-01-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Endonuclease V with DNA Reveal Initiation of Deaminated Adenine Repair.
Nat.Struct.Mol.Biol., 16, 2009
|
|
7S89
 
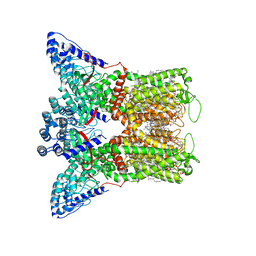 | | Open apo-state cryo-EM structure of human TRPV6 in cNW11 nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2021-09-17 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural mechanisms of TRPV6 inhibition by ruthenium red and econazole.
Nat Commun, 12, 2021
|
|
6YYJ
 
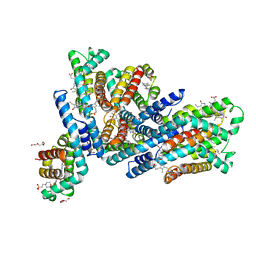 | | Crystal structure of native Phycocyanin from T. elongatus in spacegroup P21212 at 2.1 Angstroms | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(~{Z})-(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, C-phycocyanin alpha chain, ... | | Authors: | Feiler, C.G, Falke, S, Sarrou, I. | | Deposit date: | 2020-05-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | C-phycocyanin as a highly attractive model system in protein crystallography: unique crystallization properties and packing-diversity screening.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7S8C
 
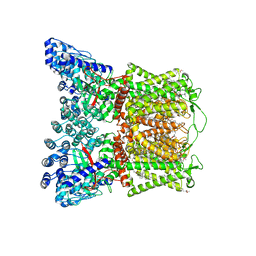 | | Cryo-EM structure of human TRPV6 in complex with inhibitor econazole | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1-[(2R)-2-[(4-chlorobenzyl)oxy]-2-(2,4-dichlorophenyl)ethyl]-1H-imidazole, CALCIUM ION, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2021-09-17 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural mechanisms of TRPV6 inhibition by ruthenium red and econazole.
Nat Commun, 12, 2021
|
|
7S88
 
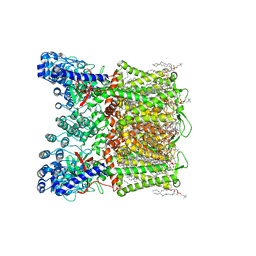 | | Open apo-state cryo-EM structure of human TRPV6 in glyco-diosgenin detergent | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2021-09-17 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural mechanisms of TRPV6 inhibition by ruthenium red and econazole.
Nat Commun, 12, 2021
|
|
8OLW
 
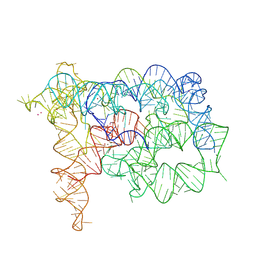 | |
7S8B
 
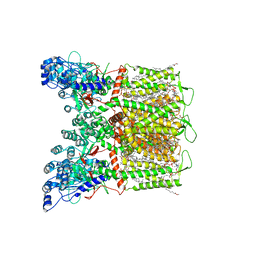 | | Cryo-EM structure of human TRPV6 in complex with channel blocker ruthenium red | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2021-09-17 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Structural mechanisms of TRPV6 inhibition by ruthenium red and econazole.
Nat Commun, 12, 2021
|
|
8OLV
 
 | | Structure of Oceanobacillus iheyensis group II intron in the presence of K+, Mg2+ and ARN25850 | | Descriptor: | 2-[2,6-bis(bromanyl)-3,4,5-tris(oxidanyl)phenyl]carbonyl-~{N}-(2-pyrrolidin-1-ylethyl)-1-benzofuran-5-carboxamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Group IIC intron, ... | | Authors: | Silvestri, I, Marcia, M. | | Deposit date: | 2023-03-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Targeting the conserved active site of splicing machines with specific and selective small molecule modulators.
Nat Commun, 15, 2024
|
|
6ZVP
 
 | | Atomic model of the EM-based structure of the full-length tyrosine hydroxylase in complex with dopamine (residues 40-497) in which the regulatory domain (residues 40-165) has been included only with the backbone atoms | | Descriptor: | FE (III) ION, L-DOPAMINE, Tyrosine 3-monooxygenase | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | Deposit date: | 2020-07-27 | | Release date: | 2021-11-17 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
6ZZU
 
 | | Partial structure of the substrate-free tyrosine hydroxylase (apo-TH). | | Descriptor: | FE (III) ION, Tyrosine 3-monooxygenase | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | Deposit date: | 2020-08-05 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
5IJI
 
 | | Fragment of nitrate/nitrite sensor histidine kinase NarQ (WT) in symmetric holo state | | Descriptor: | EICOSANE, NITRATE ION, Nitrate/nitrite sensor histidine kinase NarQ | | Authors: | Gushchin, I, Melnikov, I, Polovinkin, V, Ishchenko, A, Popov, A, Gordeliy, V. | | Deposit date: | 2016-03-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism of transmembrane signaling by sensor histidine kinases.
Science, 356, 2017
|
|
