6ILU
 
 | | Endolysin LysPBC5 CBD | | Descriptor: | 1,2-ETHANEDIOL, Lysin, SULFATE ION | | Authors: | Suh, J.Y, Ryu, K.S, Ryu, S, Lee, K.O, Kong, M.S, Bae, J.W, Kim, I.T. | | Deposit date: | 2018-10-19 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural Basis for Cell-Wall Recognition by Bacteriophage PBC5 Endolysin.
Structure, 27, 2019
|
|
8OG3
 
 | | E. coli NfsB triple mutant T41L/N71S/F124T bound to citrate | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Day, M.A, White, S.A, Hyde, E.I, Searle, P.F. | | Deposit date: | 2023-03-17 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure and Dynamics of Three Escherichia coli NfsB Nitro-Reductase Mutants Selected for Enhanced Activity with the Cancer Prodrug CB1954.
Int J Mol Sci, 24, 2023
|
|
8SGZ
 
 | | Leishmania tarentolae propionyl-CoA carboxylase (alpha-6-beta-6) | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Propionyl-coa carboxylase beta chain, putative, ... | | Authors: | Lee, J.K.J, Liu, Y.T, Hu, J.J, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2023-04-13 | | Release date: | 2023-05-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | CryoEM reveals oligomeric isomers of a multienzyme complex and assembly mechanics.
J Struct Biol X, 7, 2023
|
|
6YFI
 
 | | Bacteriophage EMS014 coat protein | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.248 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
8SGX
 
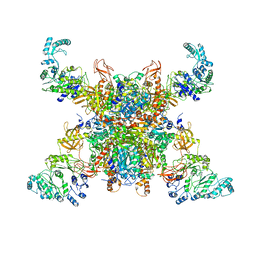 | | Leishmania tarentolae propionyl-CoA carboxylase (alpha-4-beta-6) | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Propionyl-coa carboxylase beta chain, putative, ... | | Authors: | Lee, J.K.J, Liu, Y.T, Hu, J.J, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2023-04-13 | | Release date: | 2023-05-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (10.3 Å) | | Cite: | CryoEM reveals oligomeric isomers of a multienzyme complex and assembly mechanics.
J Struct Biol X, 7, 2023
|
|
8SGY
 
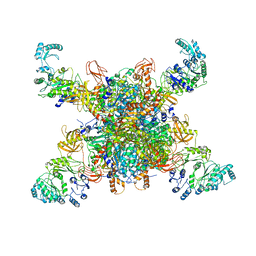 | | Leishmania tarentolae propionyl-CoA carboxylase (alpha-5-beta-6) | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Propionyl-coa carboxylase beta chain, putative, ... | | Authors: | Lee, J.K.J, Liu, Y.T, Hu, J.J, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2023-04-13 | | Release date: | 2023-05-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (8.62 Å) | | Cite: | CryoEM reveals oligomeric isomers of a multienzyme complex and assembly mechanics.
J Struct Biol X, 7, 2023
|
|
3LIO
 
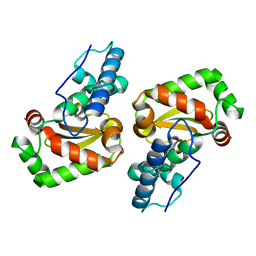 | | X-ray structure of the iron superoxide dismutase from pseudoalteromonas haloplanktis (crystal form I) | | Descriptor: | FE (III) ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, iron superoxide dismutase | | Authors: | Merlino, A, Russo Krauss, I, Rossi, B, Conte, M, Vergara, A, Sica, F. | | Deposit date: | 2010-01-25 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis.
J.Struct.Biol., 172, 2010
|
|
1QBB
 
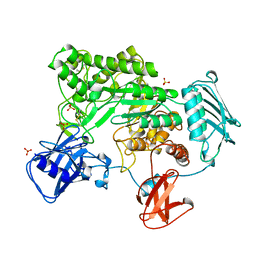 | | BACTERIAL CHITOBIASE COMPLEXED WITH CHITOBIOSE (DINAG) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-07 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
6Y7U
 
 | | Structure of Chloroflexus aggregans Cagg_3753 LOV domain C85A A95P variant (CagFbFP) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gordeliy, V, Gushchin, I. | | Deposit date: | 2020-03-02 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Effects of Proline Substitutions on the Thermostable LOV Domain from Chloroflexus aggregans
Crystals, 2020
|
|
8SO2
 
 | | Human CYP3A4 bound to a substrate | | Descriptor: | CAFFEINE, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Interaction of CYP3A4 with caffeine: First insights into multiple substrate binding.
J.Biol.Chem., 299, 2023
|
|
2ER9
 
 | | X-RAY STUDIES OF ASPARTIC PROTEINASE-STATINE INHIBITOR COMPLEXES. | | Descriptor: | ENDOTHIAPEPSIN, L363,564 | | Authors: | Cooper, J.B, Foundling, S.I, Boger, J, Blundell, T.L. | | Deposit date: | 1990-10-20 | | Release date: | 1991-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray studies of aspartic proteinase-statine inhibitor complexes.
Biochemistry, 28, 1989
|
|
6I4M
 
 | | Crystal Structure of Plasmodium berghei actin II in the Mg-ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-2, CALCIUM ION, ... | | Authors: | Kumpula, E.-P, Lopez, A.J, Tajedin, L, Han, H, Kursula, I. | | Deposit date: | 2018-11-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.873 Å) | | Cite: | Atomic view into Plasmodium actin polymerization, ATP hydrolysis, and fragmentation.
Plos Biol., 17, 2019
|
|
7V05
 
 | | Complex of Plasmodium falciparum circumsporozoite protein with 850 Fab | | Descriptor: | 850 Fab Heavy Chain, 850 Fab Light Chain, Circumsporozoite protein | | Authors: | Kucharska, I, Prieto, K, Rubinstein, J.L, Julien, J.P. | | Deposit date: | 2022-05-09 | | Release date: | 2022-11-23 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | High-density binding to Plasmodium falciparum circumsporozoite protein repeats by inhibitory antibody elicited in mouse with human immunoglobulin repertoire.
Plos Pathog., 18, 2022
|
|
1QIQ
 
 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (ACmC Fe COMPLEX) | | Descriptor: | FE (III) ION, ISOPENICILLIN N SYNTHASE, N-[N-[2-AMINO-6-OXO-HEXANOIC ACID-6-YL]CYSTEINYL]-S-METHYLCYSTEINE, ... | | Authors: | Rutledge, P.J, Clifton, I.J, Burzlaff, N.I, Roach, P.L, Adlington, R.M, Baldwin, J.E. | | Deposit date: | 1999-06-15 | | Release date: | 2000-06-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Reaction Cycle of Isopenicillin N Synthase Observed by X-Ray Diffraction.
Nature, 401, 1999
|
|
4YPA
 
 | | ASH1L SET domain Q2265A mutant in complex with S-adenosyl methionine (SAM) | | Descriptor: | Histone-lysine N-methyltransferase ASH1L, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Rogawski, D.S, Ndoj, J, Cho, H.J, Maillard, I, Grembecka, J, Cierpicki, T. | | Deposit date: | 2015-03-12 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two Loops Undergoing Concerted Dynamics Regulate the Activity of the ASH1L Histone Methyltransferase.
Biochemistry, 54, 2015
|
|
1QJF
 
 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (Monocyclic Sulfoxide - Fe COMPLEX) | | Descriptor: | 1-[(1S)-CARBOXY-2-(METHYLSULFINYL)ETHYL]-(3R)-[(5S)-5-AMINO-5-CARBOXYPENTANAMIDO]-(4R)-SULFANYLAZETIDIN-2-ONE, FE (II) ION, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Rutledge, P.J, Clifton, I.J, Burzlaff, N.I, Roach, P.L, Adlington, R.M, Baldwin, J.E. | | Deposit date: | 1999-06-23 | | Release date: | 2000-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Reaction Cycle of Isopenicillin N Synthase Observed by X-Ray Diffraction
Nature, 401, 1999
|
|
6WH2
 
 | | Structure of the C-terminal BRCT domain of human XRCC1 | | Descriptor: | X-ray repair cross complementing protein 1 variant | | Authors: | Pourfarjam, Y, Ellenberger, T, Tainer, J.A, Tomkinson, A.E, Kim, I.K. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.414 Å) | | Cite: | An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase III alpha within a flexible DNA repair complex.
Nucleic Acids Res., 49, 2021
|
|
1QJE
 
 | | Isopenicillin N synthase from Aspergillus nidulans (IP1 - Fe complex) | | Descriptor: | FE (II) ION, ISOPENICILLIN N, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Burzlaff, N.I, Clifton, I.J, Rutledge, P.J, Roach, P.L, Adlington, R.M, Baldwin, J.E. | | Deposit date: | 1999-06-23 | | Release date: | 2000-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Reaction Cycle of Isopenicillin N Synthase Observed by X-Ray Diffraction
Nature, 401, 1999
|
|
2ER6
 
 | | The structure of a synthetic pepsin inhibitor complexed with endothiapepsin. | | Descriptor: | ENDOTHIAPEPSIN, H-256 peptide | | Authors: | Cooper, J.B, Foundling, S.I, Szelke, M, Blundell, T.L. | | Deposit date: | 1990-10-13 | | Release date: | 1991-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of a synthetic pepsin inhibitor complexed with endothiapepsin
Eur.J.Biochem., 169, 1987
|
|
7UYL
 
 | | 850 Fab | | Descriptor: | 850 Fab Heavy Chain, 850 Fab Light Chain, GLYCEROL, ... | | Authors: | Kucharska, I, Prieto, K, Julien, J.P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-density binding to Plasmodium falciparum circumsporozoite protein repeats by inhibitory antibody elicited in mouse with human immunoglobulin repertoire.
Plos Pathog., 18, 2022
|
|
1NF3
 
 | | Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6 | | Descriptor: | G25K GTP-binding protein, placental isoform, MAGNESIUM ION, ... | | Authors: | Garrard, S.M, Capaldo, C.T, Gao, L, Rosen, M.K, Macara, I.G, Tomchick, D.R. | | Deposit date: | 2002-12-12 | | Release date: | 2003-03-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6
Embo J., 22, 2003
|
|
8ODU
 
 | | Chaetomium thermophilum Get1/Get2 heterotetramer in complex with a Get3 dimer (amphipol) | | Descriptor: | ATPase GET3, Protein GET2,Protein GET1, ZINC ION | | Authors: | McDowell, M.A, Wild, K, Sinning, I. | | Deposit date: | 2023-03-09 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
6WN8
 
 | | 2.70 Angstrom Resolution Crystal Structure of Uracil Phosphoribosyl Transferase from Klebsiella pneumoniae | | Descriptor: | CHLORIDE ION, SULFATE ION, Uracil phosphoribosyltransferase, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-22 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
8ODV
 
 | | Chaetomium thermophilum Get1/Get2 heterotetramer in complex with a Get3 dimer (nanodisc) | | Descriptor: | ATPase GET3, Protein GET2,Protein GET1, ZINC ION | | Authors: | McDowell, M.A, Wild, K, Sinning, I. | | Deposit date: | 2023-03-09 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
5SV0
 
 | | Structure of the ExbB/ExbD complex from E. coli at pH 7.0 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Biopolymer transport protein ExbB, CALCIUM ION | | Authors: | Celia, H, Botos, I, Lloubes, R, Buchanan, S.K, Noinaj, N. | | Deposit date: | 2016-08-04 | | Release date: | 2016-09-28 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into the role of the Ton complex in energy transduction.
Nature, 538, 2016
|
|
