8OOU
 
 | |
8C5C
 
 | |
6W7S
 
 | | Ketoreductase from module 1 of the 6-deoxyerythronolide B synthase (KR1) in complex with antibody fragment (Fab) 2G10 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2G10 (Fab heavy chain), 2G10 (Fab light chain), ... | | Authors: | Cogan, D.P, Mathews, I.I, Khosla, C. | | Deposit date: | 2020-03-19 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Antibody Probes of Module 1 of the 6-Deoxyerythronolide B Synthase Reveal an Extended Conformation During Ketoreduction.
J.Am.Chem.Soc., 142, 2020
|
|
8OP1
 
 | | Subsection of a helical nucleocapsid of the Respiratory Syncytial Virus | | Descriptor: | Nucleoprotein, RNA (5'-R(P*CP*CP*CP*CP*CP*CP*C)-3') | | Authors: | Gonnin, L, Desfosses, A, Eleouet, J.F, Galloux, M, Gutsche, I. | | Deposit date: | 2023-04-06 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural landscape of the respiratory syncytial virus nucleocapsids.
Nat Commun, 14, 2023
|
|
8CAG
 
 | | Hypoxanthine-guanine phosphoribosyltransferase from E. coli | | Descriptor: | Hypoxanthine phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Timofeev, V.I, Shevtsov, M.B, Abramchik, Y.A, Kostromina, M.A, Zayats, E.A, Kuranova, I.P, Esipov, R.S. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hypoxanthine-guanine phosphoribosyltransferase from E. coli
To Be Published
|
|
7ZOR
 
 | | Crystal structure of anti-Siglec-15 Fab | | Descriptor: | Anti-siglec15 FAB HC, Anti-siglec15 FAB LC | | Authors: | Lenza, M.P, Oyenarte, I, Jimenez-Barbero, J, Ereno-Orbea, J. | | Deposit date: | 2022-04-26 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.933 Å) | | Cite: | Structural insights into Siglec-15 reveal glycosylation dependency for its interaction with T cells through integrin CD11b.
Nat Commun, 14, 2023
|
|
1ODN
 
 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (OXYGEN-EXPOSED PRODUCT FROM ANAEROBIC AC-VINYLGLYCINE FE COMPLEX) | | Descriptor: | 6-(5-AMINO-5-CARBOXY-PENTANOYLAMINO)-3-HYDROXYMETHYL-7-OXO-4-THIA-1-AZA-BICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, FE (II) ION, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Elkins, J.M, Rutledge, P.J, Burzlaff, N.I, Clifton, I.J, Adlington, R.M, Roach, P.L, Baldwin, J.E. | | Deposit date: | 2003-02-19 | | Release date: | 2003-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Studies on the Reaction of Isopenicillin N Synthase with an Unsaturated Substrate Analogue
Org.Biomol.Chem., 1, 2003
|
|
1YN6
 
 | | Crystal structure of a mouse MHC class I protein, H2-Db, in complex with a peptide from the influenza A acid polymerase | | Descriptor: | 10-mer peptide from RNA-directed RNA polymerase subunit P2, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Turner, S.J, Kedzierska, K, Komodromou, H, La Gruta, N.L, Dunstone, M.A, Webb, A.I, Webby, R, Walden, H, Xie, W, McCluskey, J, Purcell, A.W, Rossjohn, J, Doherty, P.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lack of prominent peptide-major histocompatibility complex features limits repertoire diversity in virus-specific CD8+ T cell populations
Nat.Immunol., 6, 2005
|
|
3MDT
 
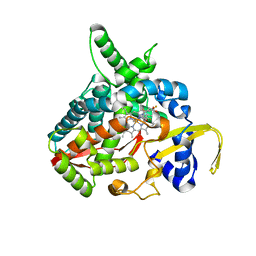 | | Voriconazole complex of Cytochrome P450 46A1 | | Descriptor: | Cholesterol 24-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE, Voriconazole | | Authors: | Mast, N, Charvet, C, Pikuleva, I, Stout, C.D. | | Deposit date: | 2010-03-30 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of drug binding to CYP46A1, an enzyme that controls cholesterol turnover in the brain.
J.Biol.Chem., 285, 2010
|
|
6CHT
 
 | | HNF4alpha in complex with the corepressor EBP1 fragment | | Descriptor: | Hepatocyte nuclear factor 4-alpha, LAURIC ACID, Proliferation-associated protein 2G4 | | Authors: | Chi, Y.I, Singh, P, Lee, I.K. | | Deposit date: | 2018-02-22 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.174 Å) | | Cite: | ErbB3-binding protein 1 (EBP1) represses HNF4 alpha-mediated transcription and insulin secretion in pancreatic beta-cells.
J.Biol.Chem., 294, 2019
|
|
6N5A
 
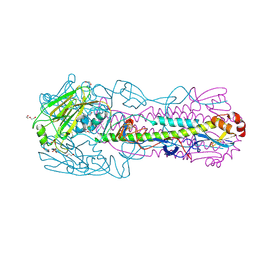 | |
1ICN
 
 | | ESCHERICHIA COLI-DERIVED RAT INTESTINAL FATTY ACID BINDING PROTEIN WITH BOUND MYRISTATE AT 1.5 A RESOLUTION AND I-FABPARG106-->GLN WITH BOUND OLEATE AT 1.74 A RESOLUTION | | Descriptor: | INTESTINAL FATTY ACID BINDING PROTEIN, OLEIC ACID | | Authors: | Eads, J.C, Sacchettini, J.C, Kromminga, A, Gordon, J.I. | | Deposit date: | 1993-09-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Escherichia coli-derived rat intestinal fatty acid binding protein with bound myristate at 1.5 A resolution and I-FABPArg106-->Gln with bound oleate at 1.74 A resolution.
J.Biol.Chem., 268, 1993
|
|
3MQI
 
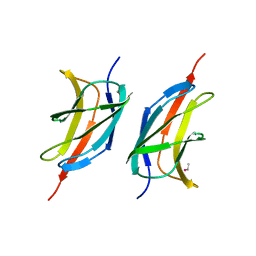 | | Human early B-cell factor 1 (EBF1) IPT/TIG domain | | Descriptor: | ETHYL MERCURY ION, Transcription factor COE1, trimethylamine oxide | | Authors: | Siponen, M.I, Lehtio, L, Arrowsmith, C.H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schueler, H, Schutz, P, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Berglund, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-28 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Determination of Functional Domains in Early B-cell Factor (EBF) Family of Transcription Factors Reveals Similarities to Rel DNA-binding Proteins and a Novel Dimerization Motif.
J.Biol.Chem., 285, 2010
|
|
1ICM
 
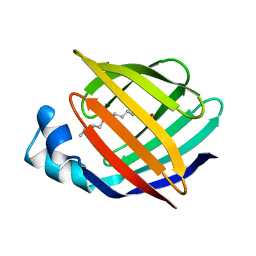 | | ESCHERICHIA COLI-DERIVED RAT INTESTINAL FATTY ACID BINDING PROTEIN WITH BOUND MYRISTATE AT 1.5 A RESOLUTION AND I-FABPARG106-->GLN WITH BOUND OLEATE AT 1.74 A RESOLUTION | | Descriptor: | INTESTINAL FATTY ACID BINDING PROTEIN, MYRISTIC ACID | | Authors: | Eads, J.C, Sacchettini, J.C, Kromminga, A, Gordon, J.I. | | Deposit date: | 1993-09-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Escherichia coli-derived rat intestinal fatty acid binding protein with bound myristate at 1.5 A resolution and I-FABPArg106-->Gln with bound oleate at 1.74 A resolution.
J.Biol.Chem., 268, 1993
|
|
8T1B
 
 | | Cryo-EM structure of full-length human TRPV4 in apo state | | Descriptor: | (2R)-2-{[(4-O-hexopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-4-{[(25R)-5beta,14beta,17beta-spirostan-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, SODIUM ION, Transient receptor potential cation channel subfamily V member 4/Enhanced green fluorescent protein chimera, ... | | Authors: | Nadezhdin, K.D, Talyzina, I.A, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
8T1F
 
 | | Cryo-EM structure of full-length human TRPV4 in complex with antagonist HC-067047 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-methyl-1-[3-(morpholin-4-yl)propyl]-5-phenyl-N-[3-(trifluoromethyl)phenyl]-1H-pyrrole-3-carboxamide, Transient receptor potential cation channel subfamily V member 4/Enhanced green fluorescent protein chimera | | Authors: | Talyzina, I.A, Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
3MDV
 
 | | Clotrimazole complex of Cytochrome P450 46A1 | | Descriptor: | 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, Cholesterol 24-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mast, N, Charvet, C, Pikuleva, I, Stout, C.D. | | Deposit date: | 2010-03-30 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of drug binding to CYP46A1, an enzyme that controls cholesterol turnover in the brain.
J.Biol.Chem., 285, 2010
|
|
8T1E
 
 | | Closed-state cryo-EM structure of full-length human TRPV4 in the presence of 4a-PDD | | Descriptor: | (2R)-2-{[(4-O-hexopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-4-{[(25R)-5beta,14beta,17beta-spirostan-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, SODIUM ION, Transient receptor potential cation channel subfamily V member 4,Enhanced green fluorescent protein, ... | | Authors: | Talyzina, I.A, Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
6WG1
 
 | |
1B7J
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 V20A | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
5OWP
 
 | | Crystal structure of glycopeptide "GVTSAfPDT*RPAP" in complex with scFv-SM3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 5,6-DIHYDRO-BENZO[H]CINNOLIN-3-YLAMINE, ... | | Authors: | Bermejo, I.A, Albuquerque, I.S, Somovilla, V.J, Martinez-Saez, N, Castro-Lopez, J, Garcia-Martin, F, Hinou, H, Nishimura, S, Jimenez-Barbero, J, Asensio, J.L, Avenoza, A, Busto, J.H, Hurtado-Guerrero, R, Peregrina, J.M, Bernardes, G.J, Corzana, F. | | Deposit date: | 2017-09-02 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Use of Fluoroproline in MUC1 Antigen Enables Efficient Detection of Antibodies in Patients with Prostate Cancer.
J. Am. Chem. Soc., 139, 2017
|
|
1AOO
 
 | | AG-SUBSTITUTED METALLOTHIONEIN FROM SACCHAROMYCES CEREVISIAE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | AG-METALLOTHIONEIN, SILVER ION | | Authors: | Peterson, C.W, Narula, S.S, Armitage, I.M. | | Deposit date: | 1997-07-08 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 3D solution structure of copper and silver-substituted yeast metallothioneins.
FEBS Lett., 379, 1996
|
|
5FMG
 
 | | Structure and function based design of Plasmodium-selective proteasome inhibitors | | Descriptor: | (2S)-N-[(E,2S)-1-(1H-indol-3-yl)-4-methylsulfonyl-but-3-en-2-yl]-2-[[(2S)-3-(1H-indol-3-yl)-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]-4-methyl-pentanamide, BETA3 PROTEASOME SUBUNIT, PUTATIVE, ... | | Authors: | Li, H, O'Donoghue, A.J, van der Linden, W.A, Xie, S.C, Yoo, E, Foe, I.T, Tilley, L, Craik, C.S, da Fonseca, P.C.A, Bogyo, M. | | Deposit date: | 2015-11-04 | | Release date: | 2016-03-02 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and Function Based Design of Plasmodium-Selective Proteasome Inhibitors
Nature, 530, 2016
|
|
8ONG
 
 | |
8P4R
 
 | | In situ structure average of GroEL14-GroES14 complexes in Escherichia coli cytosol obtained by cryo electron tomography | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (11.9 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
