7LP3
 
 | | Structure of Nedd4L WW3 domain | | Descriptor: | Angiomotin, E3 ubiquitin-protein ligase NEDD4-like, SULFATE ION | | Authors: | Alian, A, Alam, S.L, Thompson, T, Rheinemann, L, Sundquist, W.I. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
3D7A
 
 | |
7LP2
 
 | | Structure of Nedd4L WW3 domain | | Descriptor: | Angiomotin, E3 ubiquitin-protein ligase, GLYCEROL, ... | | Authors: | Alian, A, Alam, S.L, Thompson, T, Rheinemann, L, Sundquist, W.I. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
5Z45
 
 | | Crystal structure of prenyltransferase AmbP1 pH6.5 complexed with GSPP and cis-indolyl vinyl isonitrile | | Descriptor: | 3-[(Z)-2-isocyanoethenyl]-1H-indole, AmbP1, GERANYL S-THIOLODIPHOSPHATE, ... | | Authors: | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7LZ1
 
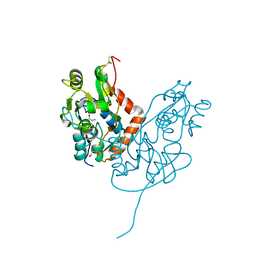 | | Structure of glutamate receptor-like channel GLR3.4 ligand-binding domain in complex with serine | | Descriptor: | GLYCEROL, Glutamate receptor 3.4, SERINE, ... | | Authors: | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | Deposit date: | 2021-03-08 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
6K90
 
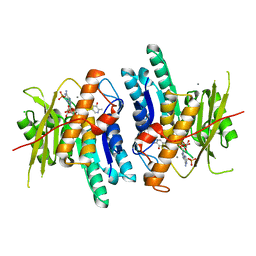 | | Pyridoxal Kinase from Leishmania donovani in complex with ADP and Pyridoxamine | | Descriptor: | 4-(AMINOMETHYL)-5-(HYDROXYMETHYL)-2-METHYLPYRIDIN-3-OL, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Are, S, Gatreddi, S, Qureshi, I.A. | | Deposit date: | 2019-06-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural attributes and substrate specificity of pyridoxal kinase from Leishmania donovani.
Int.J.Biol.Macromol., 152, 2020
|
|
2F3M
 
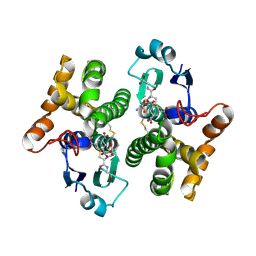 | | Structure of human GLUTATHIONE S-TRANSFERASE M1A-1A complexed with 1-(S-(GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXADIENATE ANION | | Descriptor: | 1-(S-GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXA-2,5-DIENE, Glutathione S-transferase Mu 1 | | Authors: | Patskovsky, Y, Patskovska, L, Almo, S.C, Listowsky, I. | | Deposit date: | 2005-11-21 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Transition state model and mechanism of nucleophilic aromatic substitution reactions catalyzed by human glutathione S-transferase M1a-1a.
Biochemistry, 45, 2006
|
|
6KB2
 
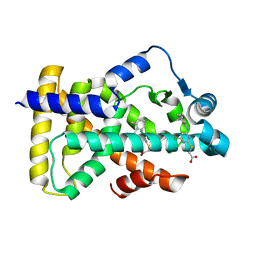 | | X-ray structure of human PPARalpha ligand binding domain-Wy14643 co-crystals obtained by soaking | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB9
 
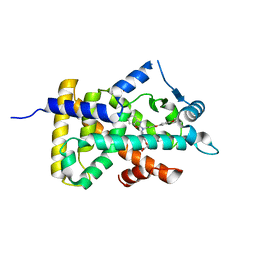 | | X-ray structure of human PPARalpha ligand binding domain-pemafibrate co-crystals obtained by cross-seeding | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
3CFC
 
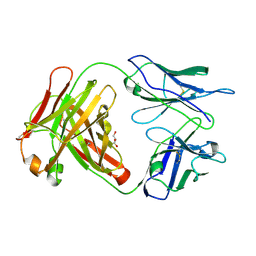 | | High-resolution structure of blue fluorescent antibody EP2-19G2 | | Descriptor: | BLUE FLUORESCENT ANTIBODY EP2-19G2-IGG2B HEAVY CHAIN, BLUE FLUORESCENT ANTIBODY EP2-19G2-KAPPA LIGHT CHAIN, GLYCEROL | | Authors: | Debler, E.W, Wilson, I.A. | | Deposit date: | 2008-03-03 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Deeply inverted electron-hole recombination in a luminescent antibody-stilbene complex.
Science, 319, 2008
|
|
3CFT
 
 | |
1XAF
 
 | | Crystal Structure of Protein of Unknown Function YfiH from Shigella flexneri 2a str. 2457T | | Descriptor: | ACETATE ION, GLYCEROL, ZINC ION, ... | | Authors: | Kim, Y, Maltseva, N, Dementieva, I, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-25 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of hypothetical protein YfiH from Shigella flexneri at 2 A resolution.
Proteins, 63, 2006
|
|
3CVY
 
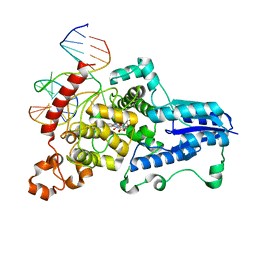 | | Drosophila melanogaster (6-4) photolyase bound to repaired ds DNA | | Descriptor: | DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DAP*DGP*DGP*DT)-3'), DNA (5'-D(*DTP*DAP*DCP*DCP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DTP*DG)-3'), FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maul, M.J, Barends, T.R.M, Glas, A.F, Cryle, M.J, Schlichting, I, Carell, T. | | Deposit date: | 2008-04-20 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and mechanism of a DNA (6-4) photolyase.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
3CN0
 
 | |
3S43
 
 | | HIV-1 protease triple mutants V32I, I47V, V82I with antiviral drug amprenavir | | Descriptor: | GLYCEROL, IODIDE ION, Protease, ... | | Authors: | Wang, Y.-F, Tie, Y.-F, Weber, I.T. | | Deposit date: | 2011-05-18 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Critical differences in HIV-1 and HIV-2 protease specificity for clinical inhibitors.
Protein Sci., 21, 2012
|
|
4PAO
 
 | | A conserved phenylalanine as relay between the 5 helix and the GDP binding region of heterotrimeric G protein | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Guanine nucleotide-binding protein G(i) subunit alpha-1, MAGNESIUM ION | | Authors: | Kaya, A.I, Lokits, A.D, Gilbert, J, Iverson, T.M, Meiler, J, Hamm, H.E. | | Deposit date: | 2014-04-09 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | A Conserved Phenylalanine as a Relay between the alpha 5 Helix and the GDP Binding Region of Heterotrimeric Gi Protein alpha Subunit.
J.Biol.Chem., 289, 2014
|
|
3COU
 
 | | Crystal structure of human Nudix motif 16 (NUDT16) | | Descriptor: | Nucleoside diphosphate-linked moiety X motif 16 | | Authors: | Tresaugues, L, Moche, M, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Svensson, L, Thorsell, A.G, Van Den Berg, S, Welin, M, Weigelt, J, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-29 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human Nudix motif 16 (NUDT16).
To be Published
|
|
1XAG
 
 | | CRYSTAL STRUCTURE OF STAPHLYOCOCCUS AUREUS 3-DEHYDROQUINATE SYNTHASE (DHQS) IN COMPLEX WITH ZN2+, NAD+ AND CARBAPHOSPHONATE | | Descriptor: | 3-dehydroquinate synthase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Nichols, C.E, Ren, J, Leslie, K, Dhaliwal, B, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-08-25 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Comparison of ligand induced conformational changes and domain closure mechanisms, between prokaryotic and eukaryotic dehydroquinate synthases.
J.Mol.Biol., 343, 2004
|
|
3Q0X
 
 | | N-terminal coiled-coil dimer domain of C. reinhardtii SAS-6 homolog Bld12p | | Descriptor: | Centriole protein | | Authors: | Kitagawa, D, Vakonakis, I, Olieric, N, Hilbert, M, Keller, D, Olieric, V, Bortfeld, M, Erat, M.C, Flueckiger, I, Goenczy, P, Steinmetz, M.O. | | Deposit date: | 2010-12-16 | | Release date: | 2011-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural basis of the 9-fold symmetry of centrioles.
Cell(Cambridge,Mass.), 144, 2011
|
|
1XAL
 
 | | CRYSTAL STRUCTURE OF STAPHLYOCOCCUS AUREUS 3-DEHYDROQUINATE SYNTHASE (DHQS) IN COMPLEX WITH ZN2+, NAD+ AND CARBAPHOSPHONATE (SOAK) | | Descriptor: | 3-dehydroquinate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION, ... | | Authors: | Nichols, C.E, Ren, J, Leslie, K, Dhaliwal, B, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-08-26 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparison of ligand induced conformational changes and domain closure mechanisms, between prokaryotic and eukaryotic dehydroquinate synthases.
J.Mol.Biol., 343, 2004
|
|
2M2F
 
 | | The membran-proximal domain of ADAM17 | | Descriptor: | Disintegrin and metalloproteinase domain-containing protein 17 | | Authors: | Duesterhoeft, S, Jung, S, Hung, C, Tholey, A, Soennichsen, F.D, Groetzinger, J, Lorenzen, I. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Membrane-proximal domain of a disintegrin and metalloprotease-17 represents the putative molecular switch of its shedding activity operated by protein-disulfide isomerase.
J.Am.Chem.Soc., 135, 2013
|
|
4PNS
 
 | | Crystal Structure of human Tankyrase 2 in complex with INH2BP. | | Descriptor: | 6-amino-5-iodo-2H-chromen-2-one, GLYCEROL, Tankyrase-2, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1X8D
 
 | | Crystal structure of E. coli YiiL protein containing L-rhamnose | | Descriptor: | Hypothetical protein yiiL, L-RHAMNOSE | | Authors: | Ryu, K.S, Kim, J.I, Cho, S.J, Park, D, Park, C, Lee, J.O, Choi, B.S. | | Deposit date: | 2004-08-18 | | Release date: | 2005-05-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the Monosaccharide Specificity of Escherichia coli Rhamnose Mutarotase
J.Mol.Biol., 349, 2005
|
|
7M1R
 
 | |
3CFM
 
 | |
