6Z1Q
 
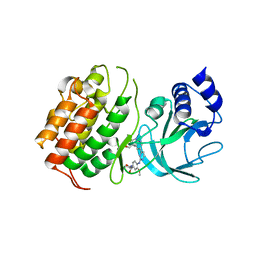 | | MAP3K14 (NIK) in complex with DesF-3R/4076 | | Descriptor: | DesF-3R/4076, Mitogen-activated protein kinase kinase kinase 14 | | Authors: | Jacoby, E, van Vlijmen, H, Querolle, O, Stansfield, I, Meerpoel, L, Versele, M, Hynd, G, Attar, R. | | Deposit date: | 2020-05-14 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | FEP+ calculations predict a stereochemical SAR switch for first-in-class indoline NIK inhibitors for multiple myeloma
Future Drug Discov, 2, 2020
|
|
6Z2M
 
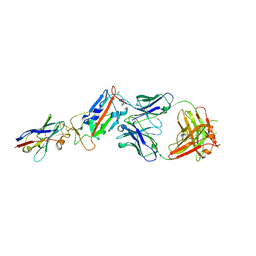 | | H11-D4, SARS-CoV-2 RBD, CR3022 ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 antibody, Spike glycoprotein, ... | | Authors: | Naismith, J.H, Ren, J, Zhou, D, Zhao, Y, Stuart, D.I. | | Deposit date: | 2020-05-17 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural characterisation of a nanobody derived from a naive library that neutralises SARS-CoV-2
To Be Published
|
|
4MZE
 
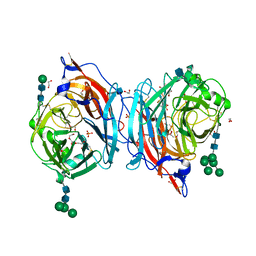 | | Crystal structure of hPIV3 hemagglutinin-neuraminidase, H552Q/Q559R mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xu, R, Wilson, I.A. | | Deposit date: | 2013-09-30 | | Release date: | 2013-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interaction between the hemagglutinin-neuraminidase and fusion glycoproteins of human parainfluenza virus type III regulates viral growth in vivo.
MBio, 4, 2013
|
|
5FOE
 
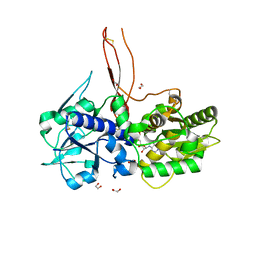 | | Crystal structure of the C. elegans Protein O-fucosyltransferase 2 (CePOFUT2) double mutant (R298K-R299K) in complex with GDP and the human TSR1 from thrombospondin 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP-fucose protein O-fucosyltransferase 2,Thrombospondin-1, ... | | Authors: | Valero-Gonzalez, J, Leonhard-Melief, C, Lira-Navarrete, E, Jimenez-Oses, G, Hernandez-Ruiz, C, Pallares, M.C, Yruela, I, Vasudevan, D, Lostao, A, Corzana, F, Takeuchi, H, Haltiwanger, R.S, Hurtado-Guerrero, R. | | Deposit date: | 2015-11-19 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A Proactive Role of Water Molecules in Acceptor Recognition of Thrombospondin Type 1 Repeats by Protein-O-Fucosyltransferase 2
Nat.Chem.Biol., 12, 2016
|
|
1ZV9
 
 | | Crystal structure analysis of a type II cohesin domain from the cellulosome of Acetivibrio cellulolyticus- SeMet derivative | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ACETIC ACID, ... | | Authors: | Noach, I, Rosenheck, S, Lamed, R, Shimon, L, Bayer, E, Frolow, F. | | Deposit date: | 2005-06-01 | | Release date: | 2006-06-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Intermodular linker flexibility revealed from crystal structures of adjacent cellulosomal cohesins of Acetivibrio cellulolyticus.
J.Mol.Biol., 391, 2009
|
|
1U2D
 
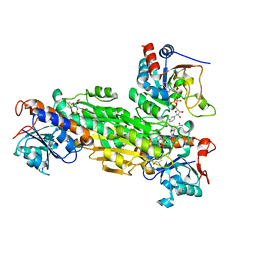 | | Structre of transhydrogenaes (dI.NADH)2(dIII.NADPH)1 asymmetric complex | | Descriptor: | GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, ... | | Authors: | Mather, O.C, Singh, A, van Boxel, G.I, White, S.A, Jackson, J.B. | | Deposit date: | 2004-07-19 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Active-site conformational changes associated with hydride transfer in proton-translocating transhydrogenase.
Biochemistry, 43, 2004
|
|
1U30
 
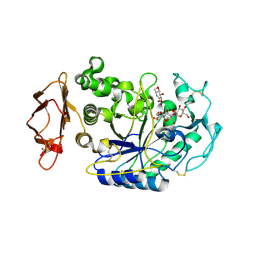 | | In situ extension as an approach for identifying novel alpha-amylase inhibitors, structure containing maltosyl-alpha (1,4)-D-gluconhydroximo-1,5-lactam | | Descriptor: | (2S,3S,4R,5R)-6-(HYDROXYAMINO)-2-(HYDROXYMETHYL)-2,3,4,5-TETRAHYDROPYRIDINE-3,4,5-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Numao, S, Li, C, Damager, I, Wrodnigg, T.M, Begum, A, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | In Situ Extension as an Approach for Identifying Novel alpha-Amylase Inhibitors.
J.Biol.Chem., 279, 2004
|
|
6ANX
 
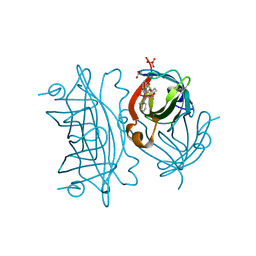 | | Peroxide Activation Regulated by Hydrogen Bonds within Artificial Cu Proteins - WT (low exposure) | | Descriptor: | ACETATE ION, SULFATE ION, Streptavidin, ... | | Authors: | Mann, S.I, Heinisch, T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2017-08-14 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Peroxide Activation Regulated by Hydrogen Bonds within Artificial Cu Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
2KL2
 
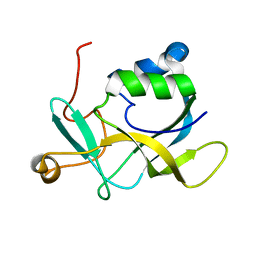 | | NMR solution structure of A2LD1 (gi:13879369) | | Descriptor: | AIG2-like domain-containing protein 1 | | Authors: | Pedrini, B, Serrano, P, Mohanty, B, Geralt, M, Herrmann, T, Wuthrich, K, Wilson, I, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures highlights conformational isomerism in protein active sites.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1U7D
 
 | |
3OB8
 
 | | Structure of the beta-galactosidase from Kluyveromyces lactis in complex with galactose | | Descriptor: | Beta-galactosidase, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Fernandez-Leiro, R, Pereira-Rodriguez, A, Becerra, M, Gonzalez-Siso, I, Cerdan, M.E, Sanz-Aparicio, J. | | Deposit date: | 2010-08-06 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of specificity in tetrameric Kluyveromyces lactis beta-galactosidase.
J.Struct.Biol., 177, 2012
|
|
5OP4
 
 | | Structure of CHK1 10-pt. mutant complex with aminopyrimidine LRRK2 inhibitor | | Descriptor: | Serine/threonine-protein kinase Chk1, [4-[[4-(ethylamino)-5-(trifluoromethyl)pyrimidin-2-yl]amino]-2-fluoranyl-5-methoxy-phenyl]-morpholin-4-yl-methanone | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OPS
 
 | | Structure of CHK1 10-pt. mutant complex with pyrrolopyridine LRRK2 inhibitor | | Descriptor: | 4-(3-hydroxyphenyl)-1~{H}-pyrrolo[2,3-b]pyridine-3-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
2NAR
 
 | | Solution structure of AVR3a_60-147 from Phytophthora infestans | | Descriptor: | Effector protein Avr3a | | Authors: | Matena, A, Bayer, P, Zhukov, I, Stanek, J, Kozminski, W, van West, P, Wawra, S. | | Deposit date: | 2016-01-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The RxLR Motif of the Host Targeting Effector AVR3a ofPhytophthora infestansIs Cleaved before Secretion.
Plant Cell, 29, 2017
|
|
6RTM
 
 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with 1-[(dimethylamino)methyl]-2-naphthol at 2 hour of soaking | | Descriptor: | 1-methylidenenaphthalen-2-one, FLAVIN-ADENINE DINUCLEOTIDE, POTASSIUM ION, ... | | Authors: | Angelucci, F, Silvestri, I, Fata, F, Williams, D.L. | | Deposit date: | 2019-05-24 | | Release date: | 2020-01-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ectopic suicide inhibition of thioredoxin glutathione reductase.
Free Radic Biol Med, 147, 2020
|
|
5THB
 
 | |
8ADB
 
 | | Viral tegument-like DUBs | | Descriptor: | CITRIC ACID, Deubiquitinating enzyme, Ubiquitin, ... | | Authors: | Erven, I, Abraham, E.T, Hermanns, T, Baumann, U, Hofmann, K. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A widely distributed family of eukaryotic and bacterial deubiquitinases related to herpesviral large tegument proteins.
Nat Commun, 13, 2022
|
|
1ZVN
 
 | | Crystal structure of chick MN-cadherin EC1 | | Descriptor: | Cadherin 1 | | Authors: | Patel, S.D, Ciatto, C, Chen, C.P, Bahna, F, Schieren, I, Rajebhosale, M, Jessell, T.M, Honig, B, Shapiro, L, Price, S.R. | | Deposit date: | 2005-06-02 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Type II cadherin ectodomain structures: implications for classical cadherin specificity.
Cell(Cambridge,Mass.), 124, 2006
|
|
6E9P
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - open form with BRD0059 bound | | Descriptor: | (2R,3S,4R)-3-(2',6'-difluoro-4'-methyl[1,1'-biphenyl]-4-yl)-4-(fluoromethyl)azetidine-2-carbonitrile, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-08-01 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.569 Å) | | Cite: | Crystal structure of tryptophan synthase from M. tuberculosis - closed form with BRD6309 bound
To be Published
|
|
2XEP
 
 | | Structural and mechanistic studies on a cephalosporin esterase from the clavulanic acid biosynthesis pathway | | Descriptor: | CHLORIDE ION, GLYCEROL, ORF12 | | Authors: | Iqbal, A, Valegard, K, Kershaw, N.J, Ivison, D, Genereux, C, Dubus, A, Andersson, I, Schofield, C.J. | | Deposit date: | 2010-05-17 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic studies of the orf12 gene product from the clavulanic acid biosynthesis pathway.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
8ADC
 
 | | Viral tegument-like DUBs | | Descriptor: | Deubiquitinating enzyme | | Authors: | Erven, I, Abraham, E.T, Hermanns, T, Baumann, U, Hofmann, K. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A widely distributed family of eukaryotic and bacterial deubiquitinases related to herpesviral large tegument proteins.
Nat Commun, 13, 2022
|
|
1KZC
 
 | | Complex of MBP-C and high-affinity linear trimannose | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN C, ... | | Authors: | Ng, K.K, Kolatkar, A.R, Park-Snyder, S, Feinberg, H, Clark, D.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Orientation of bound ligands in mannose-binding proteins. Implications for multivalent ligand recognition.
J.Biol.Chem., 277, 2002
|
|
5C6L
 
 | | Crystal Structure of Gadolinium derivative of HEWL solved using intense Free-Electron Laser radiation | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Galli, L, Barends, T.R.M, Son, S.-K, White, T.A, Barty, A, Botha, S, Boutet, S, Caleman, C, Doak, R.B, Nanao, M.H, Nass, K, Shoeman, R.L, Timneanu, N, Santra, R, Schlichting, I, Chapman, H.N. | | Deposit date: | 2015-06-23 | | Release date: | 2015-07-08 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Towards phasing using high X-ray intensity.
Iucrj, 2, 2015
|
|
2NM0
 
 | | Crystal Structure of SCO1815: a Beta-Ketoacyl-Acyl Carrier Protein Reductase from Streptomyces coelicolor A3(2) | | Descriptor: | Probable 3-oxacyl-(Acyl-carrier-protein) reductase | | Authors: | Khosla, C, Tang, Y, Lee, H.Y, Tang, Y, Kim, C.Y, Mathews, I.I. | | Deposit date: | 2006-10-20 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and functional studies on SCO1815: a beta-ketoacyl-acyl carrier protein reductase from Streptomyces coelicolor A3(2).
Biochemistry, 45, 2006
|
|
6RJI
 
 | | X-ray structure of the elongation factor P of S. aureus | | Descriptor: | Elongation factor P | | Authors: | Fatkhullin, B.F, Golubev, A.A, Gabdulkhakov, A.G, Khusainov, I.S, Validov, S.Z, Usachev, K.S, Yusupova, G, Yusupov, M.M. | | Deposit date: | 2019-04-27 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | NMR and crystallographic structural studies of the Elongation factor P from Staphylococcus aureus.
Eur.Biophys.J., 49, 2020
|
|
