8ECZ
 
 | | Bovine Fab 4C1 | | Descriptor: | 4C1 Fab heavy chain, 4C1 Fab light chain, PHOSPHATE ION | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3EIH
 
 | | Crystal structure of S.cerevisiae Vps4 in the presence of ATPgammaS | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gonciarz, M.D, Whitby, F.G, Eckert, D.M, Kieffer, C, Heroux, A, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2008-09-15 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Biochemical and structural studies of yeast vps4 oligomerization.
J.Mol.Biol., 384, 2008
|
|
8FNX
 
 | | Crystal structure of Hyaluronate lyase B from Cutibacterium acnes | | Descriptor: | GLYCEROL, Hyaluronate lyase, PHOSPHATE ION | | Authors: | Katiki, M, McNally, R, Chatterjee, A, Hajam, I.A, Liu, G.Y, Murali, R. | | Deposit date: | 2022-12-28 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional divergence of a bacterial enzyme promotes healthy or acneic skin.
Nat Commun, 14, 2023
|
|
8E4G
 
 | |
1BV4
 
 | | APO-MANNOSE-BINDING PROTEIN-C | | Descriptor: | PROTEIN (MANNOSE-BINDING PROTEIN-C) | | Authors: | Ng, K.K.-S, Weis, W.I. | | Deposit date: | 1998-09-22 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ca2+-dependent structural changes in C-type mannose-binding proteins.
Biochemistry, 37, 1998
|
|
8QQ1
 
 | | SpNOX dehydrogenase domain, mutant F397W in complex with Flavin adenine dinucleotide (FAD) | | Descriptor: | BROMIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Oxidoreductase | | Authors: | Humm, A.S, Dupeux, F, Vermot, A, Petit-Harleim, I, Fieschi, F, Marquez, J.A. | | Deposit date: | 2023-10-03 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | X-ray structure and enzymatic study of a bacterial NADPH oxidase highlight the activation mechanism of eukaryotic NOX.
Elife, 13, 2024
|
|
1YJU
 
 | | Solution structure of the apo form of the sixth soluble domain of Menkes protein | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Rosato, A, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-15 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An atomic-level investigation of the disease-causing A629P mutant of the Menkes protein, ATP7A
J.Mol.Biol., 352, 2005
|
|
1YJR
 
 | | Solution structure of the apo form of the sixth soluble domain A69P mutant of Menkes protein | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Rosato, A, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-15 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An atomic-level investigation of the disease-causing A629P mutant of the Menkes protein, ATP7A
J.Mol.Biol., 352, 2005
|
|
1BY0
 
 | |
2GGP
 
 | | Solution structure of the Atx1-Cu(I)-Ccc2a complex | | Descriptor: | COPPER (I) ION, Metal homeostasis factor ATX1, Probable copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Cantini, F, Felli, I.C, Gonnelli, L, Hadjiliadis, N, Pierattelli, R, Rosato, A, Voulgaris, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-24 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Atx1-Ccc2 complex is a metal-mediated protein-protein interaction.
Nat.Chem.Biol., 2, 2006
|
|
4UOO
 
 | |
1C08
 
 | | CRYSTAL STRUCTURE OF HYHEL-10 FV-HEN LYSOZYME COMPLEX | | Descriptor: | ANTI-HEN EGG WHITE LYSOZYME ANTIBODY (HYHEL-10), LYSOZYME | | Authors: | Shiroishi, M, Kondo, H, Matsushima, M, Tsumoto, K, Kumagai, I. | | Deposit date: | 1999-07-15 | | Release date: | 2000-07-19 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of anti-Hen egg white lysozyme antibody (HyHEL-10) Fv-antigen complex. Local structural changes in the protein antigen and water-mediated interactions of Fv-antigen and light chain-heavy chain interfaces.
J.Biol.Chem., 274, 1999
|
|
8FXI
 
 | | Cryo-EM structure of Stanieria sp. CphA2 in complex with ADPCP and 4x(beta-Asp-Arg) | | Descriptor: | 1-[(4-aminopyrimidin-5-yl)amino]-2,5-anhydro-1-deoxy-6-O-[(S)-hydroxy{[(R)-hydroxy(phosphonomethyl)phosphoryl]oxy}phosphoryl]-D-allitol, 4x(beta-Asp-Arg), MAGNESIUM ION, ... | | Authors: | Markus, L.M, Sharon, I, Strauss, M, Schmeing, T.M. | | Deposit date: | 2023-01-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and function of a hexameric cyanophycin synthetase 2.
Protein Sci., 32, 2023
|
|
1BMR
 
 | | ALPHA-LIKE TOXIN LQH III FROM SCORPION LEIURUS QUINQUESTRIATUS HEBRAEUS, NMR, 25 STRUCTURES | | Descriptor: | LQH III ALPHA-LIKE TOXIN | | Authors: | Krimm, I, Gilles, N, Sautiere, P, Stankiewicz, M, Pelhate, M, Gordon, D, Lancelin, J.-M. | | Deposit date: | 1998-07-24 | | Release date: | 1999-02-16 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structures and activity of a novel alpha-like toxin from the scorpion Leiurus quinquestriatus hebraeus.
J.Mol.Biol., 285, 1999
|
|
5HD8
 
 | |
8G2W
 
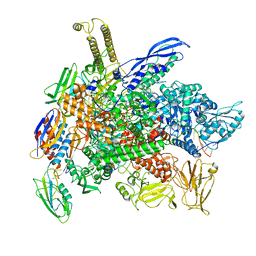 | | Cryo-EM structure of 3DVA component 2 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | Descriptor: | DNA (31-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2023-02-06 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5EH0
 
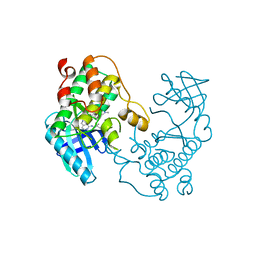 | | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach | | Descriptor: | DIMETHYL SULFOXIDE, Dual specificity protein kinase TTK, N2-(2-Methoxy-4-(1-methyl-1H-pyrazol-4-yl)phenyl)-N8-neopentylpyrido[3,4-d]pyrimidine-2,8-diamine | | Authors: | Innocenti, P, Woodward, H.L, Solanki, S, Naud, N, Westwood, I.M, Cronin, N, Hayes, A, Roberts, J, Henley, A.T, Baker, R, Faisal, A, Mak, G, Box, G, Valenti, M, De Haven Brandon, A, O'Fee, L, Saville, J, Schmitt, J, Burke, R, van Montfort, R.L.M, Raymaud, F.I, Eccles, S.A, Linardopoulos, S, Blagg, J, Hoelder, S. | | Deposit date: | 2015-10-27 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle Kinase 1 (MPS1) Using a Structure-Based Hybridization Approach.
J.Med.Chem., 59, 2016
|
|
8G1S
 
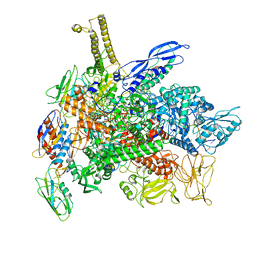 | | Cryo-EM structure of 3DVA component 1 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | Descriptor: | DNA (31-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2023-02-02 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5V8K
 
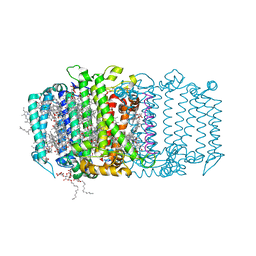 | | Homodimeric reaction center of H. modesticaldum | | Descriptor: | 1-[GLYCEROLYLPHOSPHONYL]-2-[8-(2-HEXYL-CYCLOPROPYL)-OCTANAL-1-YL]-3-[HEXADECANAL-1-YL]-GLYCEROL, 4,4'-Diaponeurosporene, 8(1)-OH-Chlorophyll aF, ... | | Authors: | Gisriel, C, Sarrou, I, Ferlez, B, Golbeck, J, Redding, K.E, Fromme, R. | | Deposit date: | 2017-03-22 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a symmetric photosynthetic reaction center-photosystem.
Science, 357, 2017
|
|
5EFO
 
 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with cytidine and cytosine at 1.63A. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-10-24 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with uridine at 2.24 A resolution
To Be Published
|
|
8SIT
 
 | |
3EJE
 
 | |
5M7O
 
 | | Crystal structure of NtrX from Brucella abortus processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | MAGNESIUM ION, Nitrogen assimilation regulatory protein | | Authors: | Cornaciu, I, Fernandez, I, Hoffmann, G, Carrica, M.C, Goldbaum, F.A, Marquez, J.A. | | Deposit date: | 2016-10-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-Dimensional Structure of Full-Length NtrX, an Unusual Member of the NtrC Family of Response Regulators.
J. Mol. Biol., 429, 2017
|
|
5EI2
 
 | | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach | | Descriptor: | Dual specificity protein kinase TTK, ~{N}-(2,4-dimethoxyphenyl)-8-(1-methylpyrazol-4-yl)pyrido[3,4-d]pyrimidin-2-amine | | Authors: | Innocenti, P, Woodward, H.L, Solanki, S, Naud, N, Westwood, I.M, Cronin, N, Hayes, A, Roberts, J, Henley, A.T, Baker, R, Faisal, A, Mak, G, Box, G, Valenti, M, De Haven Brandon, A, O'Fee, L, Saville, J, Schmitt, J, Burke, R, van Montfort, R.L.M, Raymaud, F.I, Eccles, S.A, Linardopoulos, S, Blagg, J, Hoelder, S. | | Deposit date: | 2015-10-29 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle Kinase 1 (MPS1) Using a Structure-Based Hybridization Approach.
J.Med.Chem., 59, 2016
|
|
5EQ0
 
 | | Crystal Structure of chromodomain of CBX8 in complex with inhibitor UNC3866 | | Descriptor: | Chromobox protein homolog 8, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-12 | | Release date: | 2015-12-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
