6HTY
 
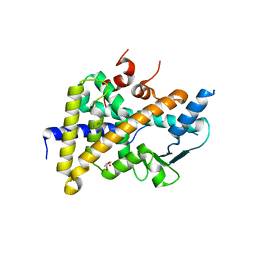 | | PXR in complex with P2X4 inhibitor compound 25 | | 分子名称: | (2~{R})-~{N}-[4-(3-chloranylphenoxy)-3-sulfamoyl-phenyl]-2-phenyl-propanamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Hillig, R.C, Puetter, V, Werner, S, Mesch, S, Laux-Biehlmann, A, Braeuer, N, Dahloef, H, Klint, J, ter Laak, A, Pook, E, Neagoe, I, Nubbemeyer, R, Schulz, S. | | 登録日 | 2018-10-05 | | 公開日 | 2019-12-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Discovery and Characterization of the Potent and Selective P2X4 InhibitorN-[4-(3-Chlorophenoxy)-3-sulfamoylphenyl]-2-phenylacetamide (BAY-1797) and Structure-Guided Amelioration of Its CYP3A4 Induction Profile.
J.Med.Chem., 62, 2019
|
|
5M4X
 
 | |
8BWO
 
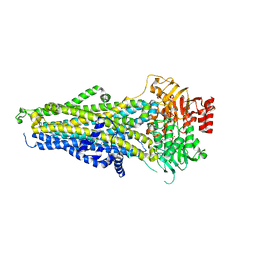 | | Cryo-EM structure of nanodisc-reconstituted human MRP4 with E1202Q mutation (outward-facing occluded conformation) | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 4, MAGNESIUM ION | | 著者 | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | 登録日 | 2022-12-07 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
8BJF
 
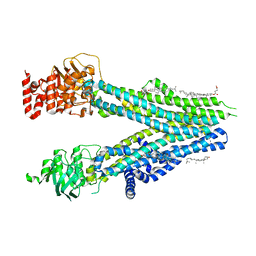 | | Cryo-EM structure of nanodisc-reconstituted wildtype human MRP4 (inward-facing conformation) | | 分子名称: | ATP-binding cassette sub-family C member 4, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE | | 著者 | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | 登録日 | 2022-11-04 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
5V5S
 
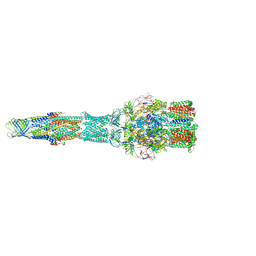 | | multi-drug efflux; membrane transport; RND superfamily; Drug resistance | | 分子名称: | Multidrug efflux pump subunit AcrA, Multidrug efflux pump subunit AcrB, Outer membrane protein TolC | | 著者 | wang, Z, fan, G, Hryc, C.F, Blaza, J.N, Serysheva, I.I, Schmid, M.F, Chiu, W, Luisi, B.F, Du, D. | | 登録日 | 2017-03-15 | | 公開日 | 2017-04-19 | | 最終更新日 | 2019-10-23 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | An allosteric transport mechanism for the AcrAB-TolC Multidrug Efflux Pump.
Elife, 6, 2017
|
|
8BWQ
 
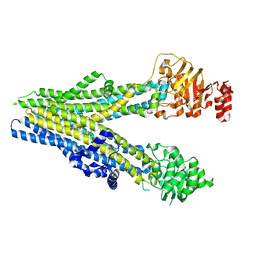 | | Cryo-EM structure of nanodisc-reconstituted wildtype human MRP4 (in complex with topotecan) | | 分子名称: | (S)-10-[(DIMETHYLAMINO)METHYL]-4-ETHYL-4,9-DIHYDROXY-1H-PYRANO[3',4':6,7]INOLIZINO[1,2-B]-QUINOLINE-3,14(4H,12H)-DIONE, ATP-binding cassette sub-family C member 4 | | 著者 | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | 登録日 | 2022-12-07 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
8BWP
 
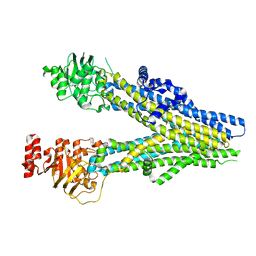 | | Cryo-EM structure of nanodisc-reconstituted wildtype human MRP4 (in complex with methotrexate) | | 分子名称: | ATP-binding cassette sub-family C member 4, METHOTREXATE | | 著者 | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | 登録日 | 2022-12-07 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
8MSI
 
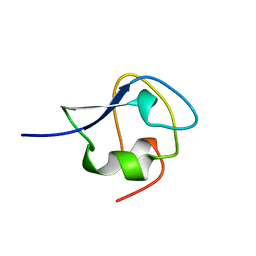 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 N14SQ44T | | 分子名称: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | 著者 | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | 登録日 | 1999-01-24 | | 公開日 | 1999-04-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
8OG3
 
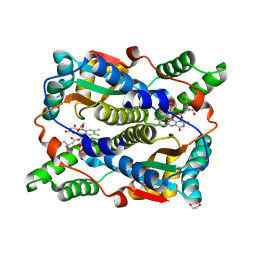 | | E. coli NfsB triple mutant T41L/N71S/F124T bound to citrate | | 分子名称: | 1,2-ETHANEDIOL, CITRIC ACID, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Day, M.A, White, S.A, Hyde, E.I, Searle, P.F. | | 登録日 | 2023-03-17 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structure and Dynamics of Three Escherichia coli NfsB Nitro-Reductase Mutants Selected for Enhanced Activity with the Cancer Prodrug CB1954.
Int J Mol Sci, 24, 2023
|
|
8OIU
 
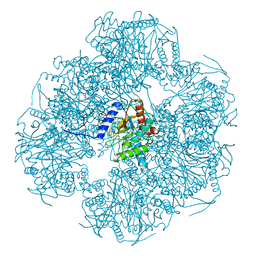 | | Cryo-EM reconstruction of the native 24-mer E2o core of the 2-oxoglutarate dehydrogenase complex of C. thermophilum at 3.35 A resolution | | 分子名称: | Dihydrolipoyllysine-residue succinyltransferase | | 著者 | Skalidis, I, Tueting, C, Kyrilis, F.L, Hamdi, F, Kastritis, P.L. | | 登録日 | 2023-03-23 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Structural analysis of an endogenous 4-megadalton succinyl-CoA-generating metabolon.
Commun Biol, 6, 2023
|
|
6XEU
 
 | | CryoEM structure of GIRK2PIP2* - G protein-gated inwardly rectifying potassium channel GIRK2 with PIP2 | | 分子名称: | G protein-activated inward rectifier potassium channel 2, POTASSIUM ION, SODIUM ION, ... | | 著者 | Mathiharan, Y.K, Glaaser, I.W, Skiniotis, G, Slesinger, P.A. | | 登録日 | 2020-06-13 | | 公開日 | 2021-09-01 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into GIRK2 channel modulation by cholesterol and PIP2
Cell Rep, 36, 2021
|
|
5S4A
 
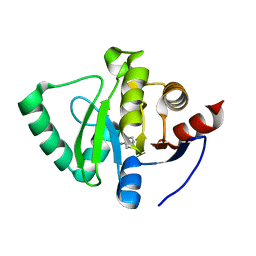 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z955123498 | | 分子名称: | 4-AMINOPYRIDINE, DIMETHYL SULFOXIDE, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.081 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7R5C
 
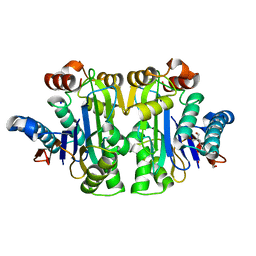 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-29 (G206C, R207S, D210L, S211V) | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Isoaspartyl peptidase, ... | | 著者 | Barciszewski, J, Imiolczyk, B, Loch, J.I, Jaskolski, M. | | 登録日 | 2022-02-10 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
1S5I
 
 | | Fab (LNKB-2) of monoclonal antibody to Human Interleukin-2, crystal structure | | 分子名称: | Fab-fragment of monoclonal antibody | | 著者 | Pletnev, V.Z, Goryacheva, E.A, Tsygannik, I.N, Nesmeyanov, V.A, Pletnev, S.V, Pangborn, W, Duax, W. | | 登録日 | 2004-01-21 | | 公開日 | 2004-05-25 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | [A new crystal form of the Fab fragment of a monoclonal antibody to human interleukin-2: the three-dimensional structure at 2.7 A resolution].
Bioorg. Khim., 30
|
|
7R1G
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-38 (R207C, D210S, S211V) | | 分子名称: | Beta-aspartyl-peptidase, Isoaspartyl peptidase, SODIUM ION | | 著者 | Loch, J.I, Kadziolka, K, Jaskolski, M. | | 登録日 | 2022-02-02 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
5JEF
 
 | | Fragment of nitrate/nitrite sensor histidine kinase NarQ (WT) in asymmetric holo state | | 分子名称: | EICOSANE, NITRATE ION, Nitrate/nitrite sensor protein NarQ | | 著者 | Gushchin, I, Melnikov, I, Polovinkin, V, Ishchenko, A, Popov, A, Gordeliy, V. | | 登録日 | 2016-04-18 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Mechanism of transmembrane signaling by sensor histidine kinases.
Science, 356, 2017
|
|
1EZT
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF FREE RGS4 BY NMR | | 分子名称: | REGULATOR OF G-PROTEIN SIGNALING 4 | | 著者 | Moy, F.J, Chanda, P.K, Cockett, M.I, Edris, W, Jones, P.G, Mason, K, Semus, S, Powers, R. | | 登録日 | 2000-05-11 | | 公開日 | 2001-01-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of free RGS4 reveals an induced conformational change upon binding Galpha.
Biochemistry, 39, 2000
|
|
1EZY
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF FREE RGS4 BY NMR | | 分子名称: | REGULATOR OF G-PROTEIN SIGNALING 4 | | 著者 | Moy, F.J, Chanda, P.K, Cockett, M.I, Edris, W, Jones, P.G, Mason, K, Semus, S, Powers, R. | | 登録日 | 2000-05-12 | | 公開日 | 2001-01-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of free RGS4 reveals an induced conformational change upon binding Galpha.
Biochemistry, 39, 2000
|
|
6XUH
 
 | | Crystal structure of human phosphoglucose isomerase in complex with inhibitor | | 分子名称: | (2R,3R,4S)-5-((2-aminoethyl)amino)-2,3,4-trihydroxy-5-oxopentyl dihydrogen phosphate, 5-PHOSPHOARABINONIC ACID, Glucose-6-phosphate isomerase | | 著者 | Li de la Sierra-Gallay, I, Ahmad, L, Plancqueel, S, van Tilbeurgh, H, Salmon, L. | | 登録日 | 2020-01-20 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Novel N-substituted 5-phosphate-d-arabinonamide derivatives as strong inhibitors of phosphoglucose isomerases: Synthesis, structure-activity relationship and crystallographic studies.
Bioorg.Chem., 102, 2020
|
|
8BBO
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.2-36 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IGH@ protein, Immunoglobulin kappa light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2022-10-14 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
5VXR
 
 | | The antigen-binding fragment of MAb24 in complex with a peptide from Hepatitis C Virus E2 epitope I (412-423) | | 分子名称: | GLYCEROL, MAb24 Variable Heavy Chain,MAb24 Variable Heavy Chain, MAb24 Variable Light Chain,MAb24 Variable Light Chain, ... | | 著者 | Hardy, J.M, Gu, J, Boo, I, Drummer, H.E, Coulibaly, F.J. | | 登録日 | 2017-05-24 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Escape of Hepatitis C Virus from Epitope I Neutralization Increases Sensitivity of Other Neutralization Epitopes.
J. Virol., 92, 2018
|
|
6HZM
 
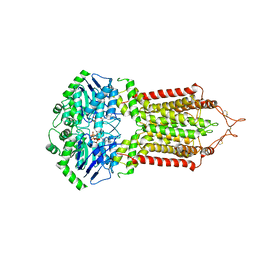 | | Cryo-EM structure of the ABCG2 E211Q mutant bound to ATP and Magnesium (alternative placement of Magnesium into the cryo-EM density) | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family G member 2, MAGNESIUM ION | | 著者 | Manolaridis, I, Jackson, S.M, Taylor, N.M.I, Kowal, J, Stahlberg, H, Locher, K.P. | | 登録日 | 2018-10-23 | | 公開日 | 2018-11-14 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.09 Å) | | 主引用文献 | Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states.
Nature, 563, 2018
|
|
7K4F
 
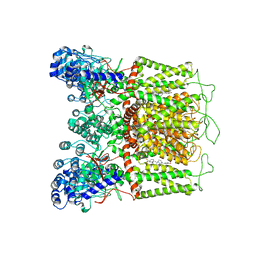 | | Cryo-EM structure of human TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor 31 | | 分子名称: | 5-[(4-{cis-4-[3-(trifluoromethyl)phenyl]cyclohexyl}piperazin-1-yl)methyl]pyridin-2(1H)-one, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | 著者 | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | 登録日 | 2020-09-15 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.75 Å) | | 主引用文献 | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
5LKS
 
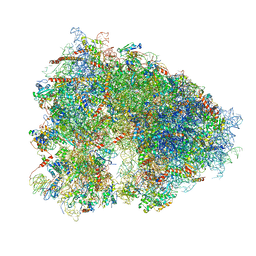 | | Structure-function insights reveal the human ribosome as a cancer target for antibiotics | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | 著者 | Myasnikov, A.G, Natchiar, S.K, Nebout, M, Hazemann, I, Imbert, V, Khatter, H, Peyron, J.-F, Klaholz, B.P. | | 登録日 | 2016-07-23 | | 公開日 | 2017-04-26 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure-function insights reveal the human ribosome as a cancer target for antibiotics.
Nat Commun, 7, 2016
|
|
6XY9
 
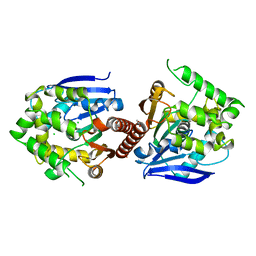 | | Crystal structure of haloalkane dehalogenase DbeA-M1 loop variant from Bradyrhizobium elkanii | | 分子名称: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase | | 著者 | Prudnikova, T, Rezacova, P, Kuta Smatanova, I, Chaloupkova, R, Damborsky, J. | | 登録日 | 2020-01-29 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural and catalytic effects of surface loop-helix transplantation within haloalkane dehalogenase family.
Comput Struct Biotechnol J, 18, 2020
|
|
