9B2H
 
 | | HIV-1 wild type protease with GRL-072-17A, a substituted tetrahydrofuran derivative based on Darunavir as P2 group | | Descriptor: | 2,5:6,9-dianhydro-1,3,7,8-tetradeoxy-4-O-({(2S,3R)-3-hydroxy-4-[(4-methoxybenzene-1-sulfonyl)(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamoyl)-L-gluco-nonitol, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2024-03-15 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Design of substituted tetrahydrofuran derivatives for HIV-1 protease inhibitors: synthesis, biological evaluation, and X-ray structural studies.
Org.Biomol.Chem., 22, 2024
|
|
6UC5
 
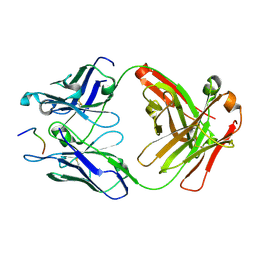 | | Fab397 in complex with NPNA peptide | | Descriptor: | Fab397 heavy chain, Fab397 light chain, NPNA peptide | | Authors: | Pholcharee, T, Oyen, D, Wilson, I.A. | | Deposit date: | 2019-09-13 | | Release date: | 2020-01-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Diverse Antibody Responses to Conserved Structural Motifs in Plasmodium falciparum Circumsporozoite Protein.
J.Mol.Biol., 432, 2020
|
|
7O7W
 
 | | Crystal structure of rsEGFP2 mutant V151L in the non-fluorescent off-state the determined by serial femtosecond crystallography at room temperature | | Descriptor: | Green fluorescent protein | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, Schlichting, I, Colletier, J.-P, Weik, M. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
7O7U
 
 | | Crystal structure of rsEGFP2 in the non-fluorescent off-state determined by serial femtosecond crystallography at room temperature | | Descriptor: | Green fluorescent protein | | Authors: | Hadjidemetriou, K, Woodhouse, J, Coquelle, N, Barends, T.R.M, Schlichting, I, Weik, M, Colletier, J.-P. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
6DPX
 
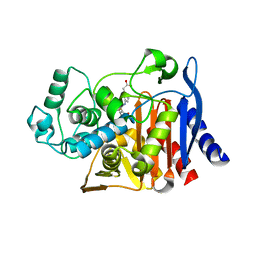 | | X-ray crystal structure of AmpC beta-lactamase with inhibitor | | Descriptor: | (3-{[(3-chloro-2-hydroxyphenyl)sulfonyl]amino}phenyl)acetic acid, Beta-lactamase | | Authors: | Singh, I. | | Deposit date: | 2018-06-09 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ultra-large library docking for discovering new chemotypes.
Nature, 566, 2019
|
|
7O7V
 
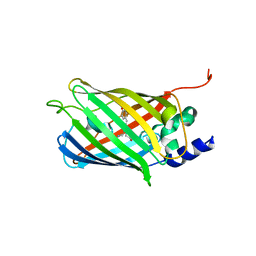 | | Crystal structure of rsEGFP2 mutant V151A in the fluorescent on-state determined by serial femtosecond crystallography at room temperature | | Descriptor: | Green fluorescent protein | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, Schlichting, I, Colletier, J.-P, Weik, M. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
7O7X
 
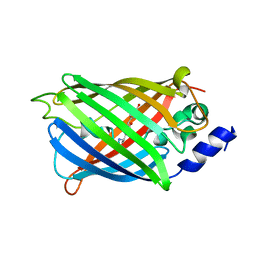 | | Crystal structure of rsEGFP2 mutant V151A in the non-fluorescent off-state determined by serial femtosecond crystallography at room temperature | | Descriptor: | Green fluorescent protein | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, Schlichting, I, Colletier, J.-P, Weik, M. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
9BC2
 
 | | Transglutaminase 2 - Open State | | Descriptor: | CHLORIDE ION, HB-225 (gluten peptidomimetic TG2 inhibitor), Protein-glutamine gamma-glutamyltransferase 2, ... | | Authors: | Mathews, I.I, Sewa, A, Khosla, C. | | Deposit date: | 2024-04-07 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and mechanistic analysis of Ca 2+ -dependent regulation of transglutaminase 2 activity using a Ca 2+ -bound intermediate state.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
2QSK
 
 | | Atomic-resolution crystal structure of the Recombinant form of Scytovirin | | Descriptor: | CHLORIDE ION, GLYCEROL, scytovirin | | Authors: | Moulaei, T, Botos, I, Ziolkowska, N.E, Dauter, Z, Wlodawer, A. | | Deposit date: | 2007-07-31 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic-resolution crystal structure of the antiviral lectin scytovirin.
Protein Sci., 16, 2007
|
|
9B69
 
 | | GluA2 flip Q in complex with TARPgamma2 at pH8, class12, structure of NTD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Flip of Glutamate receptor 2, ... | | Authors: | Nakagawa, T, Greger, I.H. | | Deposit date: | 2024-03-23 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Proton-triggered rearrangement of the AMPA receptor N-terminal domains impacts receptor kinetics and synaptic localization.
Nat.Struct.Mol.Biol., 2024
|
|
9B68
 
 | | GluA2 flip Q in complex with TARPgamma2 at pH8, class1, structure of NTD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Flip of Glutamate receptor 2, ... | | Authors: | Nakagawa, T, Greger, I.H. | | Deposit date: | 2024-03-23 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Proton-triggered rearrangement of the AMPA receptor N-terminal domains impacts receptor kinetics and synaptic localization.
Nat.Struct.Mol.Biol., 2024
|
|
7X1K
 
 | |
2FIT
 
 | | FHIT (FRAGILE HISTIDINE TRIAD PROTEIN) | | Descriptor: | FRAGILE HISTIDINE PROTEIN, SULFATE ION, beta-D-fructofuranose | | Authors: | Lima, C.D, D'Amico, K.L, Naday, I, Rosenbaum, G, Westbrook, E.M, Hendrickson, W.A. | | Deposit date: | 1997-05-17 | | Release date: | 1997-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MAD analysis of FHIT, a putative human tumor suppressor from the HIT protein family.
Structure, 5, 1997
|
|
9BC4
 
 | | Transglutaminase 2 - Intermediate State | | Descriptor: | CALCIUM ION, GLYCEROL, HB-225 (gluten peptidomimetic TG2 inhibitor), ... | | Authors: | Sewa, A.S, Mathews, I.I, Khosla, C. | | Deposit date: | 2024-04-07 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural and mechanistic analysis of Ca 2+ -dependent regulation of transglutaminase 2 activity using a Ca 2+ -bound intermediate state.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7OHE
 
 | | A self-complementary DNA dodecamer duplex contaning 5-hydroxymethylcitosine | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*CP*GP*AP*CP*GP*CP*G)-3') | | Authors: | Battistini, F, Dans, P.D, Terrazas, M, Castellazzi, C.L, Portella, G, Labrador, M, Villegas, N, Brun-Heath, I, Gonzalez, C, Orozco, M. | | Deposit date: | 2021-05-10 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Impact of the HydroxyMethylCytosine epigenetic signature on DNA structure and function.
Plos Comput.Biol., 17, 2021
|
|
7X9R
 
 | | Crystal structure of the antirepressor GmaR | | Descriptor: | Glycosyl transferase family 2 | | Authors: | Cho, S.Y, Na, H.W, Oh, H.B, Kwak, Y.M, Song, W.S, Park, S.C, Yoon, S.I. | | Deposit date: | 2022-03-16 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of flagellar motility regulation by the MogR repressor and the GmaR antirepressor in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
7OFG
 
 | |
7OHM
 
 | | A self-complementary DNA dodecamer duplex contaning 5-hydroxymethylcitosine | | Descriptor: | DNA (5'-D(*CP*GP*AP*(DH)P*GP*TP*CP*G)-3') | | Authors: | Battistini, F, Dans, P.D, Terrazas, M, Castellazzi, C.L, Portella, G, Labrador, M, Villegas, N, Brun-Heath, I, Gonzalez, C, Orozco, M. | | Deposit date: | 2021-05-11 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Impact of the HydroxyMethylCytosine epigenetic signature on DNA structure and function.
Plos Comput.Biol., 17, 2021
|
|
7OHJ
 
 | | A self-complementary DNA dodecamer duplex contaning 5-hydroxymethylcitosine | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*GP*TP*CP*G)-3') | | Authors: | Battistini, F, Dans, P.D, Terrazas, M, Castellazzi, C.L, Portella, G, Labrador, M, Villegas, N, Brun-Heath, I, Gonzalez, C, Orozco, M. | | Deposit date: | 2021-05-11 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Impact of the HydroxyMethylCytosine epigenetic signature on DNA structure and function.
Plos Comput.Biol., 17, 2021
|
|
7OGV
 
 | | A self-complementary DNA dodecamer duplex contaning 5-hydroxymethylcitosine | | Descriptor: | DNA (5'-D(*(P*GP*CP*GP*TP*(DH)P*GP*AP*CP*GP*CP*G-3') | | Authors: | Battistini, F, Dans, P.D, Terrazas, M, Castellazzi, C.L, Portella, G, Labrador, M, Villegas, N, Brun-Heath, I, Gonzalez, C, Orozco, M. | | Deposit date: | 2021-05-07 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Impact of the HydroxyMethylCytosine epigenetic signature on DNA structure and function.
Plos Comput.Biol., 17, 2021
|
|
7X9S
 
 | | Crystal structure of a complex between the antirepressor GmaR and the transcriptional repressor MogR | | Descriptor: | GmaR, Motility gene repressor MogR | | Authors: | Cho, S.Y, Na, H.W, Oh, H.B, Kwak, Y.M, Song, W.S, Park, S.C, Yoon, S.I. | | Deposit date: | 2022-03-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis of flagellar motility regulation by the MogR repressor and the GmaR antirepressor in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
2QG8
 
 | | Plasmodium yoelii acyl carrier protein synthase PY06285 with ADP bound | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Acyl Carrier Protein Synthase PY06285, MAGNESIUM ION | | Authors: | Lunin, V.V, Wernimont, A.K, Lew, J, Wasney, G, Kozieradzki, I, Vedadi, M, Bochkarev, A, Arrowsmith, C.H, Sundstrom, M, Weigelt, J, Edwards, A.E, Hui, R, Brokx, S, Altamentova, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Plasmodium yoelii acyl carrier protein synthase PY06285 with ADP bound
TO BE PUBLISHED
|
|
2QGE
 
 | |
7XHY
 
 | | Crystal structure of MerTK Kinase domain with BMS794833 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Tyrosine-protein kinase Mer, ... | | Authors: | Kim, J.H, Lee, B.I. | | Deposit date: | 2022-04-11 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | BMS794833 inhibits macrophage efferocytosis by directly binding to MERTK and inhibiting its activity.
Exp.Mol.Med., 54, 2022
|
|
8TJ8
 
 | | CRYSTAL STRUCTURE OF THE A/Moscow/10/1999(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
