1HZ8
 
 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF A CONCATEMER OF EGF-HOMOLOGY MODULES OF THE HUMAN LOW DENSITY LIPOPROTEIN RECEPTOR | | Descriptor: | CALCIUM ION, LOW DENSITY LIPOPROTEIN RECEPTOR | | Authors: | Kurniawan, N.D, Aliabadizadeh, K, Brereton, I.M, Kroon, P.A, Smith, R. | | Deposit date: | 2001-01-23 | | Release date: | 2001-08-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure and backbone dynamics of a concatemer of epidermal growth factor homology modules of the human low-density lipoprotein receptor.
J.Mol.Biol., 311, 2001
|
|
4Y0D
 
 | | Gamma-aminobutyric acid aminotransferase inactivated by (1S,3S)-3-amino-4-difluoromethylenyl-1-cyclopentanoic acid (CPP-115) | | Descriptor: | (1S)-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-3-ene-1,3-dicarboxylic acid, 4-aminobutyrate aminotransferase, mitochondrial, ... | | Authors: | Rui, W, Ruslan, S, Hyunbeom, L, Emma, H.D, Jose, I.J, Neil, K, Richard, B.S, Dali, L. | | Deposit date: | 2015-02-05 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Mechanism of inactivation of gamma-aminobutyric acid aminotransferase by (1S,3S)-3-amino-4-difluoromethylenyl-1-cyclopentanoic acid (CPP-115)
J. Am. Chem. Soc., 2015
|
|
2QG7
 
 | | Plasmodium vivax ethanolamine kinase Pv091845 | | Descriptor: | SULFATE ION, ethanolamine kinase Pv091845 | | Authors: | Lunin, V.V, Wernimont, A.K, Mulichak, A, Lew, J, Wasney, G, Senisterra, G, Kozieradzki, I, Vedadi, M, Bochkarev, A, Arrowsmith, C.H, Sundstrom, M, Weigelt, J, Edwards, A.E, Hui, R, Hills, T, Artz, J, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Plasmodium vivax ethanolamine kinase Pv091845
TO BE PUBLISHED
|
|
4I0N
 
 | | Pore forming protein | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Necrotic enteritis toxin B | | Authors: | Yan, X, Porter, C.J, Hardy, S.P, Steer, D, Smith, A.I, Quinset, N, Hughes, V, Cheung, J.K, Keyburn, A.L, Kaldhusdal, M, Moore, R.J, Bannam, T.L, Whisstock, J.C, Rood, J.I. | | Deposit date: | 2012-11-16 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analysis of the pore-forming toxin NetB from Clostridium perfringens
MBio, 4, 2013
|
|
6CEE
 
 | | Crystal structure of fragment 3-(1-Methyl-2-oxo-1,2-dihydroquinoxalin-3-yl)propionic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(4-methyl-3-oxo-3,4-dihydroquinoxalin-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Franzoni, I, Ravichandran, M, Lautens, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
1I0U
 
 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF A CONCATEMER OF EGF-HOMOLOGY MODULES OF THE HUMAN LOW DENSITY LIPOPROTEIN RECEPTOR | | Descriptor: | CALCIUM ION, LOW DENSITY LIPOPROTEIN RECEPTOR | | Authors: | Kurniawan, N.D, Aliabadizadeh, K, Brereton, I.M, Kroon, P.A, Smith, R. | | Deposit date: | 2001-01-29 | | Release date: | 2001-08-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure and backbone dynamics of a concatemer of epidermal growth factor homology modules of the human low-density lipoprotein receptor.
J.Mol.Biol., 311, 2001
|
|
5U36
 
 | | Crystal Structure Of A Mutant M. Jannashii Tyrosyl-tRNA Synthetase | | Descriptor: | Tyrosine--tRNA ligase | | Authors: | Luo, X, Fu, G, Zhu, X, Wilson, I.A, Wang, F. | | Deposit date: | 2016-12-01 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Genetically encoding phosphotyrosine and its nonhydrolyzable analog in bacteria.
Nat. Chem. Biol., 13, 2017
|
|
5U4H
 
 | | 1.05 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Acinetobacter baumannii in Covalently Bound Complex with (2R)-2-(phosphonooxy)propanoic Acid. | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, FORMIC ACID, SODIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-04 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | 1.05 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Acinetobacter baumannii in Covalently Bound Complex with (2R)-2-(phosphonooxy)propanoic Acid.
To Be Published
|
|
8C4L
 
 | |
1I5B
 
 | | STRUCTURE OF CHEA DOMAIN P4 IN COMPLEX WITH ADPNP AND MANGANESE | | Descriptor: | ACETATE ION, CHEMOTAXIS PROTEIN CHEA, MANGANESE (II) ION, ... | | Authors: | Bilwes, A.M, Quezada, C.M, Croal, L.R, Crane, B.R, Simon, M.I. | | Deposit date: | 2001-02-26 | | Release date: | 2001-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Nucleotide binding by the histidine kinase CheA.
Nat.Struct.Biol., 8, 2001
|
|
1CT8
 
 | | CATALYTIC ANTIBODY 7C8 COMPLEX | | Descriptor: | 7C8 FAB FRAGMENT; LONG CHAIN, 7C8 FAB FRAGMENT; SHORT CHAIN, SULFATE ION, ... | | Authors: | Gigant, B, Tsumuraya, T, Fujii, I, Knossow, M. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Diverse structural solutions to catalysis in a family of antibodies.
Structure Fold.Des., 7, 1999
|
|
1MSJ
 
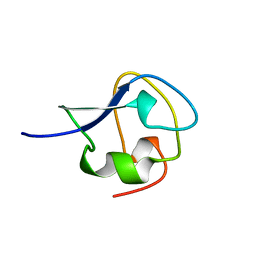 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T15V | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
8CBA
 
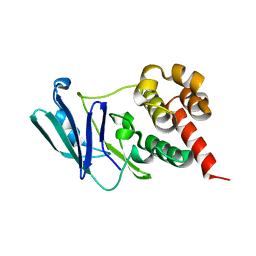 | |
8C9I
 
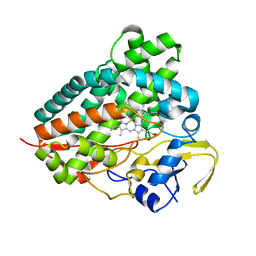 | |
5TPU
 
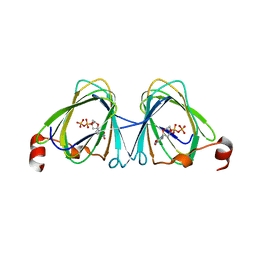 | | x-ray structure of the WlaRB TDP-quinovose 3,4-ketoisomerase from campylobacter jejuni | | Descriptor: | CHLORIDE ION, Putative uncharacterized protein, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Holden, H.M, Thoden, J.B, Li, J.Z, Riegert, A.S, Goneau, M.-F, Cunningham, A.M, Vinogradov, E, Schoenhofen, I.C, Gilbert, M. | | Deposit date: | 2016-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the dTDP-Fuc3N and dTDP-Qui3N biosynthetic pathways in Campylobacter jejuni 81116.
Glycobiology, 27, 2017
|
|
8CLO
 
 | | Zearalenone lactonase from Streptomyces coelicoflavus | | Descriptor: | 1,2-ETHANEDIOL, Hydrolase | | Authors: | Puehringer, D, Grishkovskaya, I, Mlynek, G, Kostan, J. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Bacterial Lactonases ZenA with Noncanonical Structural Features Hydrolyze the Mycotoxin Zearalenone.
Acs Catalysis, 14, 2024
|
|
1IT0
 
 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with lactose | | Descriptor: | beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, endo-1,4-beta-D-xylanase | | Authors: | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2001-12-27 | | Release date: | 2002-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
6D0P
 
 | | 1.88 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Kiryukhina, O, Endres, M, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-10 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii
To be Published
|
|
8COO
 
 | | Solution structure of Zipcode binding protein 1 (ZBP1) KH3(DD)KH4 domains in complex with N6-Methyladenosine containing RNA | | Descriptor: | Insulin-like growth factor 2 mRNA-binding protein 1, RNA_(5'-R(*(UP*CP*GP*GP*(6MZ)P*CP*U)-3') | | Authors: | Nicastro, G, Abis, G, Taylor, I.A, Ramos, A. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-07 | | Method: | SOLUTION NMR | | Cite: | Direct m6A recognition by IMP1 underlays an alternative model of target selection for non-canonical methyl-readers.
Nucleic Acids Res., 51, 2023
|
|
4XBG
 
 | | Crystal structure of human 4E10 Fab in complex with phosphatidic acid (06:0 PA): 2.73 A resolution | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 4E10 Fab heavy chain, 4E10 Fab light chain, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-16 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
3KMB
 
 | |
1J1A
 
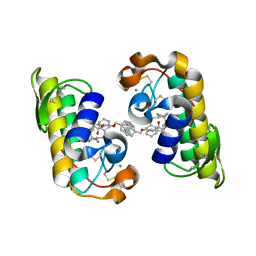 | | PANCREATIC SECRETORY PHOSPHOLIPASE A2 (IIa) WITH ANTI-INFLAMMATORY ACTIVITY | | Descriptor: | (S)-5-(4-BENZYLOXY-PHENYL)-4-(7-PHENYL-HEPTANOYLAMINO)-PENTANOIC ACID, CALCIUM ION, Phospholipase A2 | | Authors: | Hansford, K.A, Reid, R.C, Clark, C.I, Tyndall, J.D.A, Whitehouse, M.W, Guthrie, T, McGeary, R.P, Schafer, K, Martin, J.L, Fairlie, D.P. | | Deposit date: | 2002-12-03 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D-Tyrosine as a Chiral Precusor to Potent Inhibitors of Human Nonpancreatic Secretory Phospholipase A2 (IIa) with Antiinflammatory Activity
Chembiochem, 4, 2003
|
|
1J0C
 
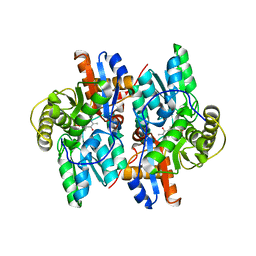 | | ACC deaminase mutated to catalytic residue | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
6CBD
 
 | |
1J31
 
 | | Crystal Structure of Hypothetical Protein PH0642 from Pyrococcus horikoshii | | Descriptor: | ACETATE ION, Hypothetical protein PH0642 | | Authors: | Sakai, N, Tajika, Y, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2003-01-16 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of hypothetical protein PH0642 from Pyrococcus horikoshii at 1.6A resolution.
Proteins, 57, 2004
|
|
