6WQW
 
 | | Thermobacillus composti GH10 xylanase | | Descriptor: | Beta-xylanase, beta-D-xylopyranose | | Authors: | Briganti, L, Polikarpov, I. | | Deposit date: | 2020-04-29 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Transformation of xylan into value-added biocommodities using Thermobacillus composti GH10 xylanase.
Carbohydr Polym, 247, 2020
|
|
7X0D
 
 | | Crystal structure of phospholipase A1, CaPLA1 | | Descriptor: | Phospholipase A1, SULFATE ION | | Authors: | Heo, Y, Lee, I, Moon, S, Lee, W. | | Deposit date: | 2022-02-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39725161 Å) | | Cite: | Crystal Structures of the Plant Phospholipase A1 Proteins Reveal a Unique Dimerization Domain.
Molecules, 27, 2022
|
|
6QII
 
 | | Xenon derivatization of the F420-reducing [NiFe] hydrogenase complex from Methanosarcina barkeri | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Coenzyme F420 hydrogenase subunit alpha, Coenzyme F420 hydrogenase subunit beta, ... | | Authors: | Ilina, Y, Lorent, C, Katz, S, Jeoung, J.H, Shima, S, Horch, M, Zebger, I, Dobbek, H. | | Deposit date: | 2019-01-19 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | X-ray Crystallography and Vibrational Spectroscopy Reveal the Key Determinants of Biocatalytic Dihydrogen Cycling by [NiFe] Hydrogenases.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6TX0
 
 | | Crystal structure of tetrameric human D137N-SAMHD1 (residues 109-626) with XTP, dAMPNPP and Mg | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Arnold, L.H, Purkiss, A.G, Kelly, G, Taylor, I.A. | | Deposit date: | 2020-01-13 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis.
Nat Commun, 11, 2020
|
|
2WWH
 
 | | Plasmodium falciparum thymidylate kinase in complex with AP5dT | | Descriptor: | P1-(5'-ADENOSYL)P5-(5'-THYMIDYL)PENTAPHOSPHATE, SODIUM ION, THYMIDILATE KINASE, ... | | Authors: | Whittingham, J.L, Carrero-Lerida, J, Brannigan, J.A, Ruiz-Perez, L.M, Silva, A.P.G, Fogg, M.J, Wilkinson, A.J, Gilbert, I.H, Wilson, K.S, Gonzalez-Pacanowska, D. | | Deposit date: | 2009-10-23 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Efficient Phosphorylation of Aztmp and Dgmp by Plasmodium Falciparum Type I Thymidylate Kinase.
Biochem.J., 428, 2010
|
|
5L3Y
 
 | | Designed Artificial Cupredoxins | | Descriptor: | Streptavidin, [CuII(biot-et-dpea)]2+ | | Authors: | Mann, S.I, Heinisch, T, Weitz, A.C, Hendrich, M.R, Ward, T.R, Borovik, A.S. | | Deposit date: | 2016-05-24 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Modular Artificial Cupredoxins.
J.Am.Chem.Soc., 138, 2016
|
|
8AYY
 
 | | Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) in complex with GSH and Pleconaril | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, Capsid protein, VP0, ... | | Authors: | Bahar, M.W, Fry, E.E, Stuart, D.I. | | Deposit date: | 2022-09-04 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A conserved glutathione binding site in poliovirus is a target for antivirals and vaccine stabilisation.
Commun Biol, 5, 2022
|
|
8AYX
 
 | | Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) in complex with GSH and GPP3 | | Descriptor: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, Capsid protein, VP0, ... | | Authors: | Bahar, M.W, Fry, E.E, Stuart, D.I. | | Deposit date: | 2022-09-04 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A conserved glutathione binding site in poliovirus is a target for antivirals and vaccine stabilisation.
Commun Biol, 5, 2022
|
|
8AYZ
 
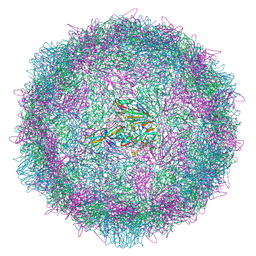 | | Poliovirus type 2 (strain MEF-1) virus-like particle in complex with capsid binder compound 17 | | Descriptor: | 4-[[4-[1,3-bis(oxidanylidene)isoindol-2-yl]phenyl]sulfonylamino]benzoic acid, Capsid protein, VP0, ... | | Authors: | Bahar, M.W, Fry, E.E, Stuart, D.I. | | Deposit date: | 2022-09-04 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (1.88 Å) | | Cite: | A conserved glutathione binding site in poliovirus is a target for antivirals and vaccine stabilisation.
Commun Biol, 5, 2022
|
|
4A6G
 
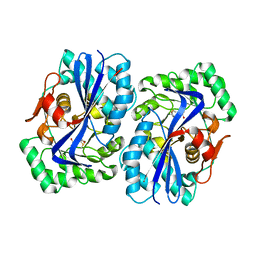 | | N-acyl amino acid racemase from Amycalotopsis sp. Ts-1-60: G291D- F323Y mutant in complex with N-acetyl methionine | | Descriptor: | MAGNESIUM ION, N-ACETYLMETHIONINE, N-ACYLAMINO ACID RACEMASE | | Authors: | Baxter, S, Royer, S, Grogan, G, Holt-Tiffin, K.E, Taylor, I.N, Fotheringham, I.G, Campopiano, D.J. | | Deposit date: | 2011-11-02 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | An Improved Racemase/Acylase Biotransformation for the Preparation of Enantiomerically Pure Amino Acids.
J.Am.Chem.Soc., 134, 2012
|
|
6WPN
 
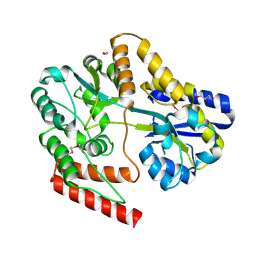 | | Crystal structure of a putative oligosaccharide periplasmic-binding protein from Synechococcus sp. MITs9220 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Substrate-binding protein | | Authors: | Ford, B.A, Michie, K.A, Paulsen, I.T, Mabbutt, B.C, Shah, B.S. | | Deposit date: | 2020-04-27 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Novel functional insights into a modified sugar-binding protein from Synechococcus MITS9220.
Sci Rep, 12, 2022
|
|
5HW5
 
 | |
6UAP
 
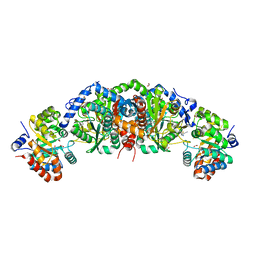 | | Crystal structure of tryptophan synthase from M. tuberculosis - open form with BRD6309 bound | | Descriptor: | (2R,3S,4R)-3-(4'-chloro-2',6'-difluoro[1,1'-biphenyl]-4-yl)-4-(fluoromethyl)azetidine-2-carbonitrile, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-11 | | Release date: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (2.745 Å) | | Cite: | Crystal structure of tryptophan synthase from M. tuberculosis - open form with BRD6309 bound
To be Published
|
|
6QGT
 
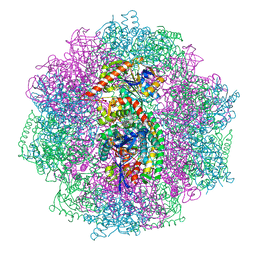 | | The carbon monoxide inhibition of F420-reducing [NiFe] hydrogenase complex from Methanosarcina barkeri | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (R,R)-2,3-BUTANEDIOL, Coenzyme F420 hydrogenase subunit beta, ... | | Authors: | Ilina, Y, Lorent, C, Katz, S, Jeoung, J.H, Shima, S, Horch, M, Zebger, I, Dobbek, H. | | Deposit date: | 2019-01-12 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | X-ray Crystallography and Vibrational Spectroscopy Reveal the Key Determinants of Biocatalytic Dihydrogen Cycling by [NiFe] Hydrogenases.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6QH8
 
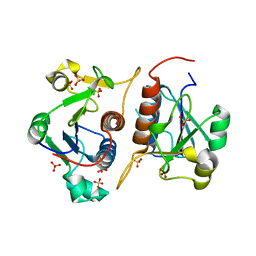 | |
6QGL
 
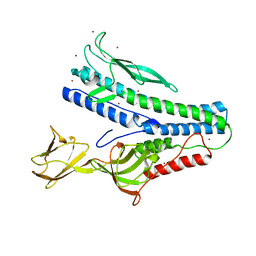 | | Crystal structure of VP5 from Haloarchaeal pleomorphic virus 6 | | Descriptor: | BROMIDE ION, VP5 | | Authors: | El Omari, K, Walter, T.S, Harlos, K, Grimes, J.M, Stuart, D.I, Roine, E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | The structure of a prokaryotic viral envelope protein expands the landscape of membrane fusion proteins.
Nat Commun, 10, 2019
|
|
5KJQ
 
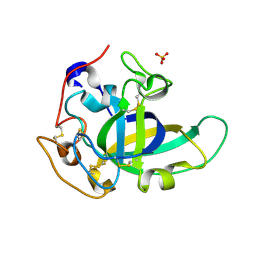 | | X-ray structure of PcCel45A in complex with cellobiose expressed in Aspergillus nidullans | | Descriptor: | Endoglucanase V-like protein, SULFATE ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Godoy, A.S, Ramia, M.P, Camilo, C.M, Polikarpov, I. | | Deposit date: | 2016-06-20 | | Release date: | 2017-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Structure, computational and biochemical analysis of PcCel45A endoglucanase from Phanerochaete chrysosporium and catalytic mechanisms of GH45 subfamily C members.
Sci Rep, 8, 2018
|
|
3PP7
 
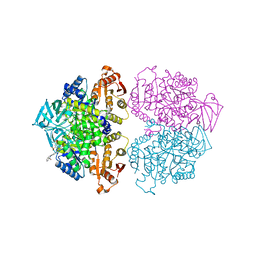 | | Crystal structure of Leishmania mexicana pyruvate kinase in complex with the drug suramin, an inhibitor of glycolysis. | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, GLYCEROL, POTASSIUM ION, ... | | Authors: | Morgan, H.P, Auld, D.S, McNae, I.W, Nowicki, M.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2010-11-24 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The trypanocidal drug suramin and other trypan blue mimetics are inhibitors of pyruvate kinases and bind to the adenosine site.
J.Biol.Chem., 286, 2011
|
|
5DF8
 
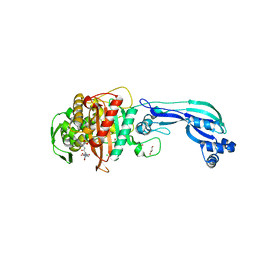 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH CEFOPERAZONE | | Descriptor: | (2R,4R,5R)-2-[(1R)-1-{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-(4-hydroxyphenyl)acetyl]amino}-2-oxoethyl]-5-methyl-1,3-thiazinane-4-carboxylic acid, CHLORIDE ION, Cell division protein, ... | | Authors: | Ren, J, Nettleship, J.E, Males, A, Stuart, D.I, Owens, R.J. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of penicillin-binding protein 3 in complexes with azlocillin and cefoperazone in both acylated and deacylated forms.
Febs Lett., 590, 2016
|
|
6UG6
 
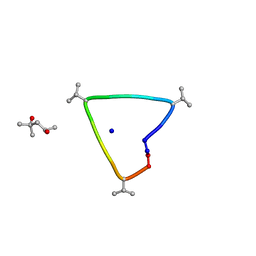 | | C3 symmetric peptide design number 1, Sporty, crystal form 2 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, C3-1, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
3GBN
 
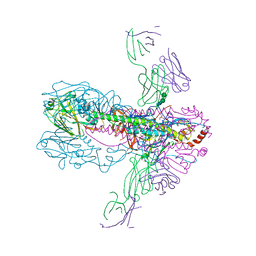 | | Crystal Structure of Fab CR6261 in Complex with the 1918 H1N1 influenza virus hemagglutinin | | Descriptor: | 1,2-ETHANEDIOL, 2-ETHOXYETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ekiert, D.C, Elsliger, M.A, Wilson, I.A. | | Deposit date: | 2009-02-20 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antibody recognition of a highly conserved influenza virus epitope.
Science, 324, 2009
|
|
6QGR
 
 | | The F420-reducing [NiFe] hydrogenase complex from Methanosarcina barkeri at the Nia-S state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (R,R)-2,3-BUTANEDIOL, Coenzyme F420 hydrogenase subunit alpha, ... | | Authors: | Ilina, Y, Lorent, C, Katz, S, Jeoung, J.H, Shima, S, Horch, M, Zebger, I, Dobbek, H. | | Deposit date: | 2019-01-12 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | X-ray Crystallography and Vibrational Spectroscopy Reveal the Key Determinants of Biocatalytic Dihydrogen Cycling by [NiFe] Hydrogenases.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6UIY
 
 | | Artificial Iron Proteins: Modelling the Active Sites in Non-Heme Dioxygenases | | Descriptor: | ACETATE ION, Streptavidin, {5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-N-(2-{[(pyridin-2-yl)methyl][(pyridin-2-yl-kappaN)methyl]amino-kappaN}ethyl)pentanamide}iron(2+) | | Authors: | Miller, K.R, Paretsky, J.D, Follmer, A.H, Heinisch, T, Mittra, K, Gul, S, Kim, I.-S, Fuller, F.D, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bhowmick, A, Sauter, N.K, Kern, J, Yano, J, Green, M.T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2019-10-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Artificial Iron Proteins: Modeling the Active Sites in Non-Heme Dioxygenases.
Inorg.Chem., 59, 2020
|
|
5ZIY
 
 | | Crystal structure of Bacillus cereus FlgL | | Descriptor: | Flagellar hook-associated protein 3, ZINC ION | | Authors: | Hong, H.J, Kim, T.H, Song, W.S, Yoon, S.I. | | Deposit date: | 2018-03-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of FlgL and its implications for flagellar assembly
Sci Rep, 8, 2018
|
|
5CSY
 
 | | Disproportionating enzyme 1 from Arabidopsis - acarbose soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
