5MSY
 
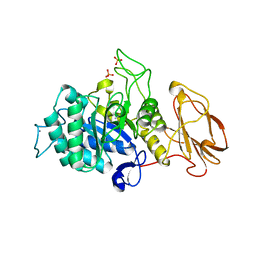 | | Glycoside hydrolase BT_1012 | | Descriptor: | AMMONIA, Glycoside hydrolase, PHOSPHATE ION | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2017-01-06 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
2O70
 
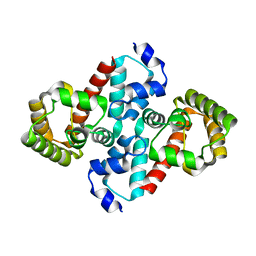 | | Structure of OHCU decarboxylase from zebrafish | | Descriptor: | OHCU decarboxylase | | Authors: | Cendron, L, Berni, R, Folli, C, Ramazzina, I, Percudani, R, Zanotti, G. | | Deposit date: | 2006-12-09 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase provides insights into the mechanism of uric acid degradation.
J.Biol.Chem., 282, 2007
|
|
1TZP
 
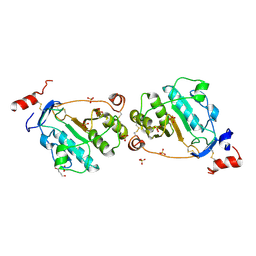 | | MEPA, inactive form without ZN in P21 | | Descriptor: | 1,4-BUTANEDIOL, Penicillin-insensitive murein endopeptidase, SULFATE ION | | Authors: | Marcyjaniak, M, Odintsov, S.G, Sabala, I, Bochtler, M. | | Deposit date: | 2004-07-11 | | Release date: | 2004-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Peptidoglycan amidase MepA is a LAS metallopeptidase
J.Biol.Chem., 279, 2004
|
|
7AC4
 
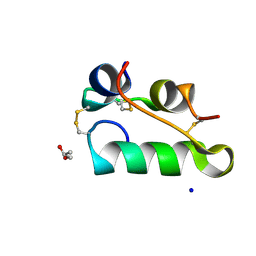 | | Structure of insulin collected by rotation serial crystallography on a COC membrane at a synchrotron source | | Descriptor: | Insulin, R-1,2-PROPANEDIOL, SODIUM ION | | Authors: | Martiel, I, Padeste, C, Karpik, A, Huang, C.Y, Vera, L, Wang, M, Marsh, M. | | Deposit date: | 2020-09-09 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Versatile microporous polymer-based supports for serial macromolecular crystallography.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1K5W
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE SYNAPTOTAGMIN 1 C2B-DOMAIN: SYNAPTOTAGMIN 1 AS A PHOSPHOLIPID BINDING MACHINE | | Descriptor: | CALCIUM ION, Synaptotagmin I | | Authors: | Fernandez, I, Arac, D, Ubach, J, Gerber, S.H, Shin, O, Gao, Y, Anderson, R.G.W, Sudhof, T.C, Rizo, J. | | Deposit date: | 2001-10-12 | | Release date: | 2002-01-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the synaptotagmin 1 C2B-domain: synaptotagmin 1 as a phospholipid binding machine.
Neuron, 32, 2001
|
|
7AC3
 
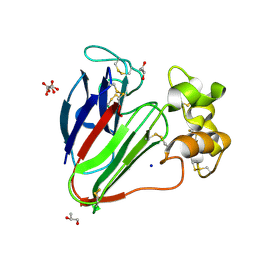 | | Structure of thaumatin collected by rotation serial crystallography on a COC membrane at a synchrotron source | | Descriptor: | L(+)-TARTARIC ACID, S-1,2-PROPANEDIOL, SODIUM ION, ... | | Authors: | Martiel, I, Padeste, C, Karpik, A, Huang, C.Y, Vera, L, Wang, M, Marsh, M. | | Deposit date: | 2020-09-09 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Versatile microporous polymer-based supports for serial macromolecular crystallography.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4GTE
 
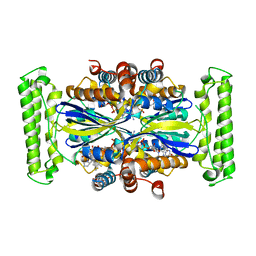 | | T. Maritima FDTS (E144R mutant) with FAD and Folate | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-({4-[(6aR)-3-amino-1-oxo-1,2,5,6,6a,7-hexahydroimidazo[1,5-f]pteridin-8(9H)-yl]phenyl}carbonyl)-L-glutamic acid, Thymidylate synthase thyX | | Authors: | Mathews, I.I, Lesley, S.A, Kohen, A, Prabhakar, A. | | Deposit date: | 2012-08-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Folate binding site of flavin-dependent thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7RYP
 
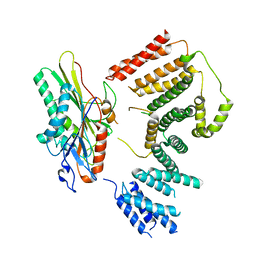 | | Cryo-EM structure of KIFBP:KIF15 | | Descriptor: | KIF-binding protein, Kinesin-like protein KIF15 | | Authors: | Solon, A.L, Tan, Z, Schutt, K.L, Jepsen, L, Haynes, S.E, Nesvizhskii, A.I, Sept, D, Stumpff, J, Ohi, R, Cianfrocco, M.A. | | Deposit date: | 2021-08-25 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Kinesin-binding protein remodels the kinesin motor to prevent microtubule binding.
Sci Adv, 7, 2021
|
|
5MEF
 
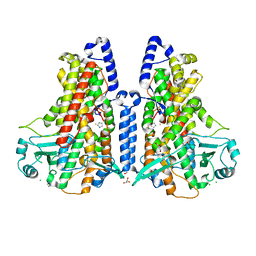 | | Cyanothece lipoxygenase 2 (CspLOX2) variant - L304F | | Descriptor: | Arachidonate 15-lipoxygenase, CHLORIDE ION, FE (III) ION, ... | | Authors: | Newie, J, Neumann, P, Werner, M, Mata, R.A, Ficner, R, Feussner, I. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.357 Å) | | Cite: | Lipoxygenase 2 from Cyanothece sp. controls dioxygen insertion by steric shielding and substrate fixation.
Sci Rep, 7, 2017
|
|
7ABD
 
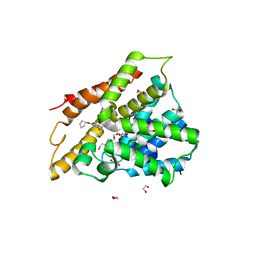 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-768 | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-cyclopentyloxy-4-methoxy-phenyl)-4,4-dimethyl-1~{H}-pyrazol-5-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2020-09-07 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-768
To be published
|
|
5MEX
 
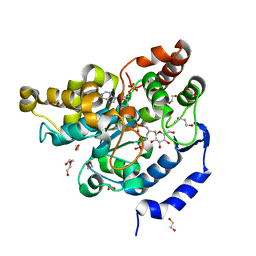 | | Sulphotransferase-18 from Arabidopsis thaliana in complex with 3'-phosphoadenosine 5'-phosphate (PAP)and sinigrin | | Descriptor: | 1,2-ETHANEDIOL, 1,3-BUTANEDIOL, 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hirschmann, F, Krause, F, Baruch, P, Chizhov, I, Mueller, J.W, Manstein, D.J, Papenbrock, J, Fedorov, R. | | Deposit date: | 2016-11-16 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and biochemical studies of sulphotransferase 18 from Arabidopsis thaliana explain its substrate specificity and reaction mechanism.
Sci Rep, 7, 2017
|
|
6DXH
 
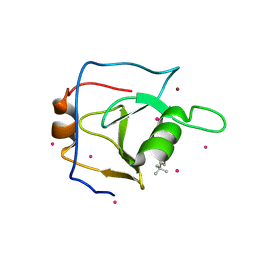 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with 4-(4-tert-butylphenyl)-4-oxobutanoate | | Descriptor: | 4-(4-tert-butylphenyl)-4-oxobutanoic acid, UNKNOWN ATOM OR ION, Ubiquitin carboxyl-terminal hydrolase 5, ... | | Authors: | Harding, R.J, Mann, M.K, Ravichandran, M, Ferreira de Freitas, R, Franzoni, I, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-28 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Small Molecule Antagonists of the USP5 Zinc Finger Ubiquitin-Binding Domain.
J.Med.Chem., 62, 2019
|
|
5MRA
 
 | | human SCBD (sorcin calcium binding domain) in complex with doxorubicin | | Descriptor: | DIMETHYL SULFOXIDE, DOXORUBICIN, MAGNESIUM ION, ... | | Authors: | Ilari, A, Fiorillo, A, Colotti, G, Genovese, I. | | Deposit date: | 2016-12-22 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.74 Å) | | Cite: | Binding of doxorubicin to Sorcin impairs cell death and increases drug resistance in cancer cells.
Cell Death Dis, 8, 2017
|
|
6DNP
 
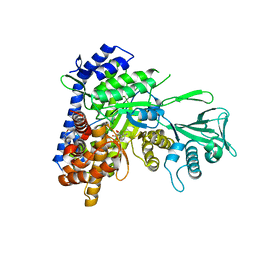 | | Crystal structure of Mycobacterium tuberculosis malate synthase in complex with 2-F-3-Methyl-6-F-phenyldiketoacid | | Descriptor: | (2Z)-4-(2,6-difluoro-3-methylphenyl)-2-hydroxy-4-oxobut-2-enoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Krieger, I.V, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC), Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.711 Å) | | Cite: | Anion-pi Interactions in Computer-Aided Drug Design: Modeling the Inhibition of Malate Synthase by Phenyl-Diketo Acids.
J Chem Inf Model, 58, 2018
|
|
6RU9
 
 | |
1JXF
 
 | | SOLUTION STRUCTURE OF REDUCED CU(I) PLASTOCYANIN FROM SYNECHOCYSTIS PCC6803 | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Bertini, I, Bryant, D.A, Ciurli, S, Dikiy, A, Fernandez, C.O, Luchinat, C, Safarov, N, Vila, A.J, Zhao, J. | | Deposit date: | 2001-09-07 | | Release date: | 2001-09-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics of plastocyanin in both oxidation states. Solution structure of the reduced form and comparison with the oxidized state.
J.Biol.Chem., 276, 2001
|
|
6Z06
 
 | | Crystal structure of Puumala virus Gc in complex with Fab 4G2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope polyprotein, Fab 4G2 Heavy chain, ... | | Authors: | Rissanen, I.R, Stass, R, Krumm, S.A, Seow, J, Hulswit, R.J.G, Paesen, G.C, Hepojoki, J, Vapalahti, O, Lundkvist, A, Reynard, O, Volchkov, V, Doores, K.J, Huiskonen, J.T, Bowden, T.A. | | Deposit date: | 2020-05-07 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular rationale for antibody-mediated targeting of the hantavirus fusion glycoprotein.
Elife, 9, 2020
|
|
1NVG
 
 | | N249Y MUTANT OF THE ALCOHOL DEHYDROGENASE FROM THE ARCHAEON SULFOLOBUS SOLFATARICUS-TETRAGONAL CRYSTAL FORM | | Descriptor: | NAD-dependent alcohol dehydrogenase, ZINC ION | | Authors: | Esposito, L, Bruno, I, Sica, F, Raia, C.A, Giordano, A, Rossi, M, Mazzarella, L, Zagari, A. | | Deposit date: | 2003-02-03 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural study of a single-point mutant of Sulfolobus solfataricus alcohol dehydrogenase with enhanced activity
Febs Lett., 539, 2003
|
|
2O3W
 
 | | Crystal Structure of the Homo sapiens Cytoplasmic Ribosomal Decoding Site in presence of paromomycin | | Descriptor: | PAROMOMYCIN, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*CP*UP*CP*CP*GP*GP*AP*AP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Hainrichson, M, Nudelman, I, Shallom-Shezifi, D, Baasov, T, Westhof, E. | | Deposit date: | 2006-12-02 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Differential Selectivity of Natural and Synthetic Aminoglycosides towards the Eukaryotic and Prokaryotic Decoding A Sites.
Chembiochem, 8, 2007
|
|
7BOH
 
 | | Complete Bacterial 30S ribosomal subunit assembly complex state E (+RbfA)(Consensus Refinement) | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
2O73
 
 | | Structure of OHCU decarboxylase in complex with allantoin | | Descriptor: | 1-(2,5-DIOXO-2,5-DIHYDRO-1H-IMIDAZOL-4-YL)UREA, OHCU decarboxylase | | Authors: | Cendron, L, Berni, R, Folli, C, Ramazzina, I, Percudani, R, Zanotti, G. | | Deposit date: | 2006-12-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase provides insights into the mechanism of uric acid degradation.
J.Biol.Chem., 282, 2007
|
|
6YUX
 
 | | Crystal structure of Malus domestica Double Bond Reductase (MdDBR) ternary complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ... | | Authors: | Caliandro, R, Polsinelli, I, Demitri, N, Benini, S. | | Deposit date: | 2020-04-27 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The structural and functional characterization of Malus domestica double bond reductase MdDBR provides insights towards the identification of its substrates.
Int.J.Biol.Macromol., 171, 2021
|
|
5OMB
 
 | | Crystal structure of K. lactis Ddc2 N-terminus in complex with S. cerevisiae Rfa1 N-OB domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA damage checkpoint protein LCD1, ... | | Authors: | Deshpande, I, Seeber, A, Shimada, K, Keusch, J.J, Gut, H, Gasser, S.M. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-25 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Basis of Mec1-Ddc2-RPA Assembly and Activation on Single-Stranded DNA at Sites of Damage.
Mol. Cell, 68, 2017
|
|
3ZTS
 
 | | Hexagonal form P6122 of the Aquifex aeolicus nucleoside diphosphate kinase (FINAL STAGE OF RADIATION DAMAGE) | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Boissier, F, Georgescauld, F, Moynie, L, Dupuy, J.-W, Sarger, C, Podar, M, Lascu, I, Giraud, M.-F, Dautant, A. | | Deposit date: | 2011-07-12 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Inter-Subunit Disulphide Bridge Stabilizes the Tetrameric Nucleoside Diphosphate Kinase of Aquifex Aeolicus
Proteins, 80, 2012
|
|
3ZTR
 
 | | Hexagonal form P6122 of the Aquifex aeolicus nucleoside diphosphate kinase (FIRST STAGE OF RADIATION DAMAGE) | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Boissier, F, Georgescauld, F, Moynie, L, Dupuy, J.-W, Sarger, C, Podar, M, Lascu, I, Giraud, M.-F, Dautant, A. | | Deposit date: | 2011-07-12 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Inter-Subunit Disulphide Bridge Stabilizes the Tetrameric Nucleoside Diphosphate Kinase of Aquifex Aeolicus
Proteins, 80, 2012
|
|
