6Z0X
 
 | |
6Z0Y
 
 | |
8IXQ
 
 | |
7QOX
 
 | | Factor XI and Plasma Kallikrein apple domain structures reveals different kininogen bound complexes | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Li, C, Awital, B, Wong, S, Dreveny, I, Meijers, J, Emsley, J. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Plasma kallikrein structure reveals apple domain disc rotated conformation compared to factor XI.
J Thromb Haemost, 17, 2019
|
|
1VYQ
 
 | | Novel inhibitors of Plasmodium Falciparum dUTPase provide a platform for anti-malarial drug design | | Descriptor: | 2,3-DEOXY-3-FLUORO-5-O-TRITYLURIDINE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Whittingham, J.L, Leal, I, Kasinathan, G, Nguyen, C, Bell, E, Jones, A.F, Berry, C, Benito, A, Turkenburg, J.P, Dodson, E.J, Ruiz Perez, L.M, Wilkinson, A.J, Johansson, N.G, Brun, R, Gilbert, I.H, Gonzalez Pacanowska, D, Wilson, K.S. | | Deposit date: | 2004-05-05 | | Release date: | 2005-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dutpase as a Platform for Antimalarial Drug Design: Structural Basis for the Selectivity of a Class of Nucleoside Inhibitors.
Structure, 13, 2005
|
|
7QOT
 
 | | Factor XI and Plasma Kallikrein apple domain structures reveals different kininogen bound complexes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XIa heavy chain, Kininogen-1 light chain, ... | | Authors: | Li, C, Awital, B, Wong, S, Dreveny, I, Meijers, J, Emsley, J. | | Deposit date: | 2021-12-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Plasma kallikrein structure reveals apple domain disc rotated conformation compared to factor XI.
J Thromb Haemost, 17, 2019
|
|
6XYV
 
 | |
7QUZ
 
 | | Crystal structure of the SeMet octameric C-terminal Big_2-CBM56 domains from Paenibacillus illinoisensis (Bacillus circulans IAM1165) beta-1,3-glucanase H | | Descriptor: | Beta-1,3-glucanase bglH, CHLORIDE ION, GLYCEROL | | Authors: | Najmudin, S, Venditto, I, Fontes, C.M.G.A, Bule, P. | | Deposit date: | 2022-01-19 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.156 Å) | | Cite: | Structural and biochemical characterization of C-terminal Big_2-CBM56 domains of Bacillus circulans IAM1165 beta-1,3-glucanase H and Paenibacillus sp CBM56
To be published
|
|
7QCB
 
 | | Williams-Beuren syndrome related methyltransferase WBSCR27 in complex with SAH | | Descriptor: | Methyltransferase-like 27, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Polshakov, V.I, Mariasina, S.S, Chang, C.-F, Efimov, S.V. | | Deposit date: | 2021-11-23 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Williams-Beuren Syndrome Related Methyltransferase WBSCR27: From Structure to Possible Function.
Front Mol Biosci, 9, 2022
|
|
1RJF
 
 | | Structure of PPM1, a leucine carboxy methyltransferase involved in the regulation of protein phosphatase 2A activity | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, carboxy methyl transferase for protein phosphatase 2A catalytic subunit | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Sorel, I, de La Sierra-Gallay, I.L, Collinet, B, Graille, M, Blondeau, K, Bettache, N, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2003-11-19 | | Release date: | 2003-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of protein phosphatase methyltransferase 1 (PPM1), a leucine carboxyl methyltransferase involved in the regulation of protein phosphatase 2A activity
J.Biol.Chem., 279, 2004
|
|
7QAC
 
 | | The T2 structure of polycrystalline cubic human insulin | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Karavassili, F, Triandafillidis, D.P, Valmas, A, Spiliopoulou, M, Fili, S, Kontou, P, Bowler, M.W, Von Dreele, R.B, Fitch, A, Margiolaki, I. | | Deposit date: | 2021-11-16 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | POWDER DIFFRACTION (2.29 Å) | | Cite: | The T 2 structure of polycrystalline cubic human insulin.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6ZQQ
 
 | | Structure of the Pmt3-MIR domain with bound ligands | | Descriptor: | GLYCEROL, PMT3 isoform 1 | | Authors: | Wild, K, Chiapparino, A, Hackmann, Y, Mortensen, S, Sinning, I. | | Deposit date: | 2020-07-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional implications of MIR domains in protein O -mannosylation.
Elife, 9, 2020
|
|
5MFA
 
 | | Crystal structure of human promyeloperoxidase (proMPO) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grishkovskaya, I, Furtmueller, P.G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-11-17 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of human promyeloperoxidase (proMPO) and the role of the propeptide in processing and maturation.
J. Biol. Chem., 292, 2017
|
|
6ZRD
 
 | |
6ZSI
 
 | | The mechanism of activation of the actin binding protein EHBP1 by Rab8 family members. | | Descriptor: | DI(HYDROXYETHYL)ETHER, EH domain-binding protein 1, MAGNESIUM ION, ... | | Authors: | Rai, A, Bleimling, N, Vetter, I.R, Goody, R.S. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | The mechanism of activation of the actin binding protein EHBP1 by Rab8 family members.
Nat Commun, 11, 2020
|
|
6ZSJ
 
 | | The mechanism of activation of the actin binding protein EHBP1 by Rab8 family members. | | Descriptor: | EH domain-binding protein 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Rai, A, Bleimling, N, Vetter, I.R, Goody, R.S. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mechanism of activation of the actin binding protein EHBP1 by Rab8 family members.
Nat Commun, 11, 2020
|
|
7RLR
 
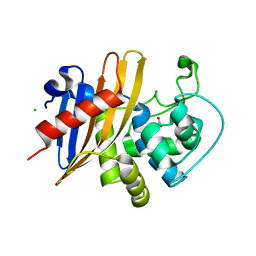 | | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beta-lactamase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-26 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
7RL8
 
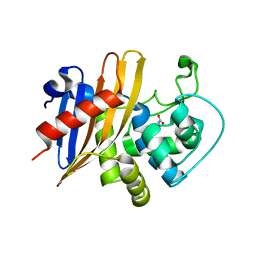 | | Crystal Structure of C79A Mutant of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-23 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of C79A Mutant of Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
6XRZ
 
 | | The 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | Descriptor: | Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | Authors: | Zhang, K, Zheludev, I, Hagey, R, Wu, M, Haslecker, R, Hou, Y, Kretsch, R, Pintilie, G, Rangan, R, Kladwang, W, Li, S, Pham, E, Souibgui, C, Baric, R, Sheahan, T, Souza, V, Glenn, J, Chiu, W, Das, R. | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Cryo-electron Microscopy and Exploratory Antisense Targeting of the 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome.
Biorxiv, 2020
|
|
6ZQP
 
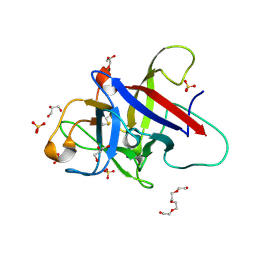 | | Structure of the Pmt2-MIR domain with bound ligands | | Descriptor: | GLYCEROL, PMT2 isoform 1, SULFATE ION, ... | | Authors: | Wild, K, Chiapparino, A, Hackmann, Y, Mortensen, S, Sinning, I. | | Deposit date: | 2020-07-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional implications of MIR domains in protein O -mannosylation.
Elife, 9, 2020
|
|
7RAS
 
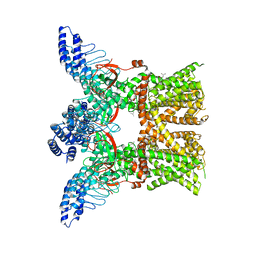 | | Structure of TRPV3 in complex with osthole | | Descriptor: | 7-methoxy-8-(3-methylbut-2-enyl)chromen-2-one, Transient receptor potential cation channel subfamily V member 3 | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2021-07-02 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural mechanism of TRPV3 channel inhibition by the plant-derived coumarin osthole.
Embo Rep., 22, 2021
|
|
6ZQV
 
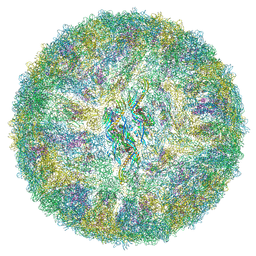 | | Cryo-EM structure of mature Spondweni virus | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein | | Authors: | Renner, M, Dejnirattisai, W, Carrique, L, Serna Martin, I, Karia, D, Ilca, S.L, Ho, S.F, Kotecha, A, Keown, J.R, Mongkolsapaya, J, Screaton, G.R, Grimes, J.M. | | Deposit date: | 2020-07-10 | | Release date: | 2021-01-20 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Flavivirus maturation leads to the formation of an occupied lipid pocket in the surface glycoproteins.
Nat Commun, 12, 2021
|
|
6ZRC
 
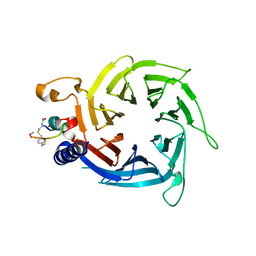 | |
7RAU
 
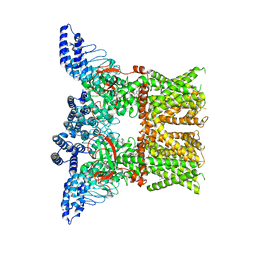 | | Structure of TRPV3 in complex with osthole | | Descriptor: | 7-methoxy-8-(3-methylbut-2-enyl)chromen-2-one, Transient receptor potential cation channel subfamily V member 3 | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2021-07-02 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural mechanism of TRPV3 channel inhibition by the plant-derived coumarin osthole.
Embo Rep., 22, 2021
|
|
7REI
 
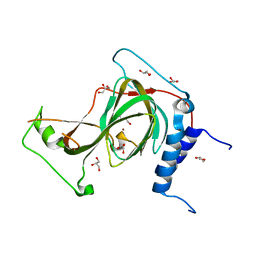 | | The crystal structure of nickel bound human ADO C18S C239S variant | | Descriptor: | 2-aminoethanethiol dioxygenase, GLYCEROL, NICKEL (II) ION | | Authors: | Wang, Y, Shin, I, Li, J, Liu, A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of human cysteamine dioxygenase provides a structural rationale for its function as an oxygen sensor.
J.Biol.Chem., 297, 2021
|
|
