7QQ8
 
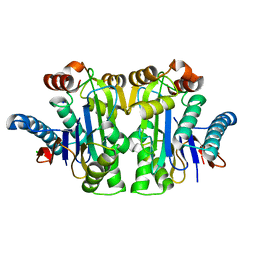 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-8 (G206Y, R207Q, D210P, S211T) | | Descriptor: | Beta-aspartyl-peptidase, CHLORIDE ION, SODIUM ION | | Authors: | Loch, J.I, Kadziolka, K, Jaskolski, M. | | Deposit date: | 2022-01-06 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6WVW
 
 | | Crystal structure of the R59P-SNAP25 containing SNARE complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Synaptosomal-associated protein 25, ... | | Authors: | Zhou, Q, White, K.I, Brunger, A.T. | | Deposit date: | 2020-05-07 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Role of Aberrant Spontaneous Neurotransmission in SNAP25-Associated Encephalopathies.
Neuron, 109, 2021
|
|
1QIQ
 
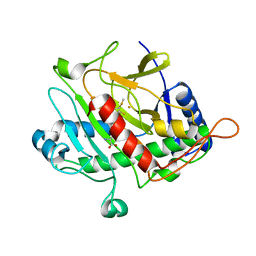 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (ACmC Fe COMPLEX) | | Descriptor: | FE (III) ION, ISOPENICILLIN N SYNTHASE, N-[N-[2-AMINO-6-OXO-HEXANOIC ACID-6-YL]CYSTEINYL]-S-METHYLCYSTEINE, ... | | Authors: | Rutledge, P.J, Clifton, I.J, Burzlaff, N.I, Roach, P.L, Adlington, R.M, Baldwin, J.E. | | Deposit date: | 1999-06-15 | | Release date: | 2000-06-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Reaction Cycle of Isopenicillin N Synthase Observed by X-Ray Diffraction.
Nature, 401, 1999
|
|
1QJF
 
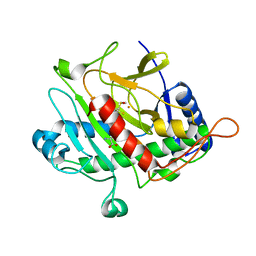 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (Monocyclic Sulfoxide - Fe COMPLEX) | | Descriptor: | 1-[(1S)-CARBOXY-2-(METHYLSULFINYL)ETHYL]-(3R)-[(5S)-5-AMINO-5-CARBOXYPENTANAMIDO]-(4R)-SULFANYLAZETIDIN-2-ONE, FE (II) ION, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Rutledge, P.J, Clifton, I.J, Burzlaff, N.I, Roach, P.L, Adlington, R.M, Baldwin, J.E. | | Deposit date: | 1999-06-23 | | Release date: | 2000-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Reaction Cycle of Isopenicillin N Synthase Observed by X-Ray Diffraction
Nature, 401, 1999
|
|
6TVV
 
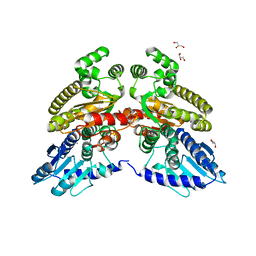 | | Crystal structure of 3'-5' RecJ exonuclease from M. Jannaschii | | Descriptor: | GLYCEROL, MANGANESE (II) ION, MjaRecJ | | Authors: | De March, M, Medagli, B, Krastanova, I, Saha, I, Pisani, F, Onesti, S. | | Deposit date: | 2020-01-10 | | Release date: | 2021-01-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of 3'-5' RecJ exonuclease from M. Jannaschii
To Be Published
|
|
1QJE
 
 | | Isopenicillin N synthase from Aspergillus nidulans (IP1 - Fe complex) | | Descriptor: | FE (II) ION, ISOPENICILLIN N, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Burzlaff, N.I, Clifton, I.J, Rutledge, P.J, Roach, P.L, Adlington, R.M, Baldwin, J.E. | | Deposit date: | 1999-06-23 | | Release date: | 2000-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Reaction Cycle of Isopenicillin N Synthase Observed by X-Ray Diffraction
Nature, 401, 1999
|
|
6WPM
 
 | | Crystal structure of a putative oligosaccharide periplasmic-binding protein from Synechococcus sp. MITs9220 in complex with zinc | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Ford, B.A, Michie, K.A, Paulsen, I.T, Mabbutt, B.C, Shah, B.S. | | Deposit date: | 2020-04-27 | | Release date: | 2021-05-12 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Novel functional insights into a modified sugar-binding protein from Synechococcus MITS9220.
Sci Rep, 12, 2022
|
|
5VXZ
 
 | | High-affinity AXL decoy receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Mathrews, I.I, Kapur, S, Kariolis, M.S, Cochran, J.R. | | Deposit date: | 2017-05-24 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibition of the GAS6/AXL pathway augments the efficacy of chemotherapies.
J. Clin. Invest., 127, 2017
|
|
6WPY
 
 | |
7QXT
 
 | | ATAD2 in complex with FragLite10 | | Descriptor: | 1,2-ETHANEDIOL, 6-bromo-1,3-dihydro-2H-indol-2-one, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-01-27 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7QUK
 
 | | ATAD2 in complex with FragLite1 | | Descriptor: | 1,2-ETHANEDIOL, 4-bromo-1H-pyrazole, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-01-18 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
5NDV
 
 | | Crystal structure of Paromomycin bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Prokhorova, I, Djumagulov, M, Urzhumtsev, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2017-03-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Aminoglycoside interactions and impacts on the eukaryotic ribosome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1LEG
 
 | | Crystal Structure of H-2Kb bound to the dEV8 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-2-MICROGLOBULIN, ... | | Authors: | Luz, J.G, Huang, M, Garcia, K.C, Rudolph, M.G, Apostolopoulos, V, Teyton, L, Wilson, I.A. | | Deposit date: | 2002-04-09 | | Release date: | 2002-06-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural comparison of allogeneic and syngeneic T cell receptor-peptide-major histocompatibility complex complexes: a buried alloreactive mutation subtly alters peptide presentation substantially increasing V(beta) Interactions.
J.Exp.Med., 195, 2002
|
|
6XK0
 
 | | Albumin-dexamethasone complex | | Descriptor: | Albumin, CITRATE ANION, DEXAMETHASONE, ... | | Authors: | Czub, M.P, Majorek, K.A, Shabalin, I.G, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2020-06-24 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular determinants of vascular transport of dexamethasone in COVID-19 therapy.
Iucrj, 7, 2020
|
|
7CDL
 
 | | holo-methanol dehydrogenase (MDH) with Cys131-Cys132 reduced from Methylococcus capsulatus (Bath) | | Descriptor: | CALCIUM ION, Methanol dehydrogenase [cytochrome c] subunit 2, Methanol dehydrogenase protein, ... | | Authors: | Chuankhayan, P, Chan, S.I, Nareddy, P.K.R, Tsai, I.K, Tsai, Y.F, Chen, K.H.-C, Yu, S.S.-F, Chen, C.J. | | Deposit date: | 2020-06-20 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of Pyrroloquinoline Quinone-Dependent Hydride Transfer Chemistry from Spectroscopic and High-Resolution X-ray Structural Studies of the Methanol Dehydrogenase from Methylococcus capsulatus (Bath).
J.Am.Chem.Soc., 143, 2021
|
|
3FOL
 
 | | Crystal structure of the Class I MHC Molecule H-2Kwm7 with a Single Self Peptide VNDIFERI | | Descriptor: | 8 residue synthetic peptide, Beta-2-microglobulin, MHC | | Authors: | Brims, D.R, Qian, J, Jarchum, I, Yamada, T, Mikesh, L, Palmieri, E, Lund, T, Hattori, M, Shabanowitz, J, Hunt, D.F, Ramagopal, U.A, Malashkevich, V.N, Almo, S.C, Nathenson, S.G, DiLorenzo, T.P. | | Deposit date: | 2008-12-30 | | Release date: | 2010-01-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Predominant occupation of the class I MHC molecule H-2Kwm7 with a single self-peptide suggests a mechanism for its diabetes-protective effect
Int.Immunol., 22, 2010
|
|
7CFX
 
 | | NAD-soaked Holo-methanol dehydrogenase (MDH) from Methylococcus capsulatus (Bath) | | Descriptor: | CALCIUM ION, Methanol dehydrogenase [cytochrome c] subunit 2, Methanol dehydrogenase protein, ... | | Authors: | Chuankhayan, P, Chan, S.I, Nareddy, P.K.R, Tsai, I.K, Tsai, Y.F, Chen, K.H.-C, Yu, S.S.-F, Chen, C.J. | | Deposit date: | 2020-06-29 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Pyrroloquinoline Quinone-Dependent Hydride Transfer Chemistry from Spectroscopic and High-Resolution X-ray Structural Studies of the Methanol Dehydrogenase from Methylococcus capsulatus (Bath).
J.Am.Chem.Soc., 143, 2021
|
|
7CED
 
 | | Apo-methanol dehydrogenase (MDH) from Methylococcus capsulatus (Bath) | | Descriptor: | Methanol dehydrogenase [cytochrome c] subunit 2, Methanol dehydrogenase protein, large subunit | | Authors: | Chuankhayan, P, Chan, S.I, Nareddy, P.K.R, Tsai, I.K, Tsai, Y.F, Chen, K.H.-C, Yu, S.S.-F, Chen, C.J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of Pyrroloquinoline Quinone-Dependent Hydride Transfer Chemistry from Spectroscopic and High-Resolution X-ray Structural Studies of the Methanol Dehydrogenase from Methylococcus capsulatus (Bath).
J.Am.Chem.Soc., 143, 2021
|
|
7CE5
 
 | | Methanol-PQQ bound methanol dehydrogenase (MDH) from Methylococcus capsulatus (Bath) | | Descriptor: | CALCIUM ION, METHANOL, Methanol dehydrogenase [cytochrome c] subunit 2, ... | | Authors: | Chuankhayan, P, Chan, S.I, Nareddy, P.K.R, Tsai, I.K, Tsai, Y.F, Chen, K.H.-C, Yu, S.S.-F, Chen, C.J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of Pyrroloquinoline Quinone-Dependent Hydride Transfer Chemistry from Spectroscopic and High-Resolution X-ray Structural Studies of the Methanol Dehydrogenase from Methylococcus capsulatus (Bath).
J.Am.Chem.Soc., 143, 2021
|
|
7CE9
 
 | | PQQ-soaked Apo-methanol dehydrogenase (MDH) from Methylococcus capsulatus (Bath) | | Descriptor: | CALCIUM ION, Methanol dehydrogenase [cytochrome c] subunit 2, Methanol dehydrogenase protein, ... | | Authors: | Chuankhayan, P, Chan, S.I, Nareddy, P.K.R, Tsai, I.K, Tsai, Y.F, Chen, K.H.-C, Yu, S.S.-F, Chen, C.J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Pyrroloquinoline Quinone-Dependent Hydride Transfer Chemistry from Spectroscopic and High-Resolution X-ray Structural Studies of the Methanol Dehydrogenase from Methylococcus capsulatus (Bath).
J.Am.Chem.Soc., 143, 2021
|
|
5NUN
 
 | | Engineered beta-lactoglobulin: variant F105L-L39A-M107F in complex with chlorpromazine (LG-LAF-CLP) | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, Beta-lactoglobulin | | Authors: | Loch, J.I, Bonarek, P, Towrzydlo, M, Lazinskia, I, Szydlowska, J, Lewinski, K. | | Deposit date: | 2017-04-30 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The engineered beta-lactoglobulin with complementarity to the chlorpromazine chiral conformers.
Int. J. Biol. Macromol., 114, 2018
|
|
6HCR
 
 | | Synthetic Self-assembling ADDomer Platform for Highly Efficient Vaccination by Genetically-encoded Multi-epitope Display | | Descriptor: | Penton protein | | Authors: | Bufton, J.C, Berger, I, Schaffitzel, C, Garzoni, F, Rabi, F.A. | | Deposit date: | 2018-08-16 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Synthetic self-assembling ADDomer platform for highly efficient vaccination by genetically encoded multiepitope display.
Sci Adv, 5, 2019
|
|
7OMT
 
 | | Crystal structure of ProMacrobody 21 with bound maltose | | Descriptor: | HEXAETHYLENE GLYCOL, MAGNESIUM ION, ProMacrobody 21, ... | | Authors: | Botte, M, Ni, D, Schenck, S, Zimmermann, I, Chami, M, Bocquet, N, Egloff, P, Bucher, D, Trabuco, M, Cheng, R.K.Y, Brunner, J.D, Seeger, M.A, Stahlberg, H, Hennig, M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation.
Nat Commun, 13, 2022
|
|
6XC7
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CC12.3 and CR3022 | | Descriptor: | CC12.3 heavy chain, CC12.3 light chain, CR3022 heavy chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
6XMP
 
 | | Structure of P5A-ATPase Spf1, Apo form | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, P5A-type ATPase | | Authors: | Park, E, Sim, S.I. | | Deposit date: | 2020-06-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase.
Science, 369, 2020
|
|
