2NQP
 
 | |
2NR0
 
 | |
2NRE
 
 | |
2GIO
 
 | |
2GIP
 
 | |
6NE0
 
 | | Structure of double-stranded target DNA engaged Csy complex from Pseudomonas aeruginosa (PA-14) | | Descriptor: | CRISPR RNA (60-MER), CRISPR target DNA (44-MER), CRISPR-associated endonuclease Cas6/Csy4, ... | | Authors: | Chowdhury, S, Rollins, M.F, Carter, J, Golden, S.M, Miettinen, H.M, Santiago-Frangos, A, Faith, D, Lawrence, M.C, Wiedenheft, B, Lander, G.C. | | Deposit date: | 2018-12-15 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Reveals a Mechanism of CRISPR-RNA-Guided Nuclease Recruitment and Anti-CRISPR Viral Mimicry.
Mol. Cell, 74, 2019
|
|
3FXE
 
 | |
5JZM
 
 | |
5JZO
 
 | |
5K4A
 
 | |
8H17
 
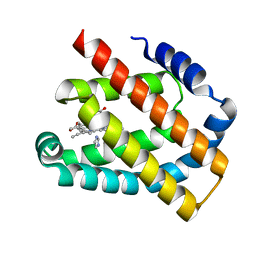 | | Crystal structure of the Globin domain of Thermosynechococcus elongatus BP-1 | | Descriptor: | IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, Tlr1989 protein | | Authors: | Mathur, S, Yadav, S.K, Pal, K.R, Kundu, S. | | Deposit date: | 2022-09-30 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A novel single sensor hemoglobin domain from the thermophilic cyanobacteria Thermosynechococcus elongatus BP-1 exhibits higher pH but lower thermal stability compared to globins from mesophilic organisms.
Int.J.Biol.Macromol., 240, 2023
|
|
3FXD
 
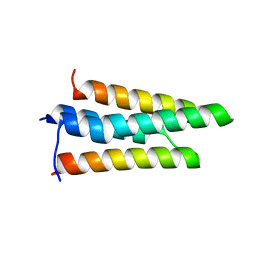 | |
5UZ9
 
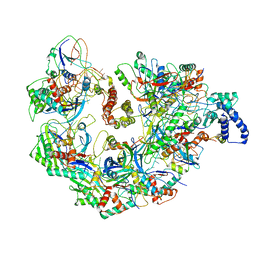 | | Cryo EM structure of anti-CRISPRs, AcrF1 and AcrF2, bound to type I-F crRNA-guided CRISPR surveillance complex | | Descriptor: | Anti-CRISPR protein 30, Anti-CRISPR protein Acr30-35, CRISPR RNA (60-MER), ... | | Authors: | Chowdhury, S, Carter, J, Rollins, M.F, Jackson, R.N, Hoffmann, C, Nosaka, L, Bondy-Denomy, J, Maxwell, K.L, Davidson, A.R, Fischer, E.R, Lander, G.C, Wiedenheft, B. | | Deposit date: | 2017-02-25 | | Release date: | 2017-04-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Reveals Mechanisms of Viral Suppressors that Intercept a CRISPR RNA-Guided Surveillance Complex.
Cell, 169, 2017
|
|
6X9K
 
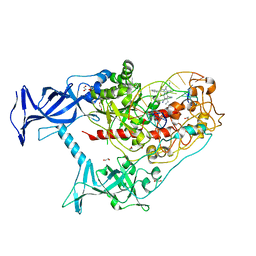 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Inhibitor GSK3685032A | | Descriptor: | (2R)-2-{[6-(4-aminopiperidin-1-yl)-3,5-dicyano-4-ethylpyridin-2-yl]sulfanyl}-2-phenylacetamide, 1,2-ETHANEDIOL, DNA (5'-D(*GP*AP*GP*GP*CP*(5CM)P*GP*CP*CP*TP*GP*C)-3'), ... | | Authors: | Pathuri, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-06-02 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia.
Nat Cancer, 2, 2021
|
|
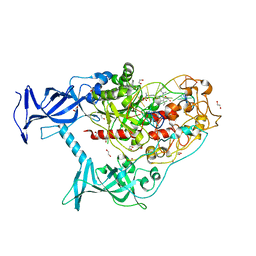
6X9J
 
 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Inhibitor GSK3830052 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*AP*GP*GP*CP*(5CM)P*GP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*G)-R(P*(PYO))-D(P*GP*GP*CP*CP*TP*C)-3'), ... | | Authors: | Pathuri, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-06-02 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia.
Nat Cancer, 2, 2021
|
|
1A80
 
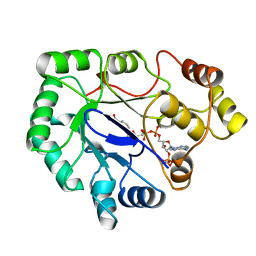 | | Native 2,5-DIKETO-D-GLUCONIC acid reductase a from CORYNBACTERIUM SP. complexed with nadph | | Descriptor: | 2,5-DIKETO-D-GLUCONIC ACID REDUCTASE A, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Khurana, S, Powers, D.B, Anderson, S, Blaber, M. | | Deposit date: | 1998-03-31 | | Release date: | 1999-03-30 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of 2,5-diketo-D-gluconic acid reductase A complexed with NADPH at 2.1-A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6X9I
 
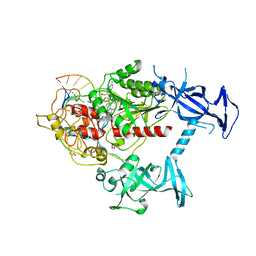 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*AP*GP*GP*CP*(5CM)P*GP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*G)-R(P*(PYO))-D(P*GP*GP*CP*CP*TP*C)-3'), ... | | Authors: | Pathuri, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-06-02 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia.
Nat Cancer, 2, 2021
|
|
1MHW
 
 | | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides | | Descriptor: | 4-biphenylacetyl-Cys-(D)Arg-Tyr-N-(2-phenylethyl) amide, Cathepsin L | | Authors: | Chowdhury, S, Sivaraman, J, Wang, J, Devanathan, G, Lachance, P, Qi, H, Menard, R, Lefebvre, J, Konishi, Y, Cygler, M, Sulea, T, Purisima, E.O. | | Deposit date: | 2002-08-21 | | Release date: | 2002-12-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides
J.Med.Chem., 45, 2002
|
|
3BC3
 
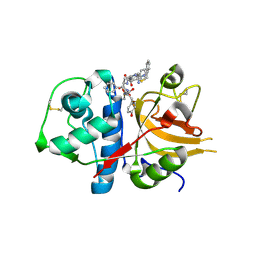 | | Exploring inhibitor binding at the S subsites of cathepsin L | | Descriptor: | Cathepsin L heavy and light chains, S-benzyl-N-(biphenyl-4-ylacetyl)-L-cysteinyl-N~5~-(diaminomethyl)-D-ornithyl-N-(2-phenylethyl)-L-tyrosinamide | | Authors: | Chowdhury, S.F, Joseph, L, Kumar, S, Tulsidas, S.R, Bhat, S, Ziomek, E, Nard, R.M, Sivaraman, J, Purisima, E.O. | | Deposit date: | 2007-11-12 | | Release date: | 2008-03-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring inhibitor binding at the S' subsites of cathepsin L
J.Med.Chem., 51, 2008
|
|
8SRO
 
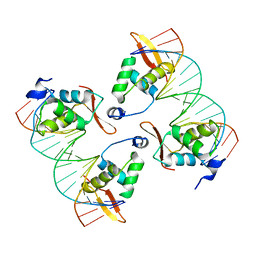 | | FoxP3 tetramer on TTTG repeats | | Descriptor: | DNA 72-mer, Forkhead box protein P3 | | Authors: | Leng, F, Hur, S. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | FOXP3 recognizes microsatellites and bridges DNA through multimerization.
Nature, 624, 2023
|
|
8SRP
 
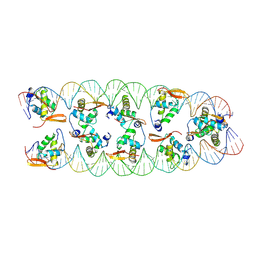 | |
4GL2
 
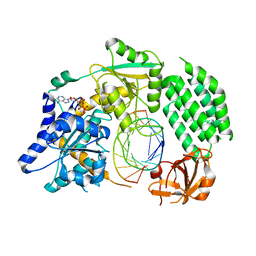 | | Structural Basis for dsRNA duplex backbone recognition by MDA5 | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RNA (5'-R(*AP*UP*CP*CP*GP*CP*GP*GP*CP*CP*CP*U)-3'), ... | | Authors: | Wu, B, Hur, S. | | Deposit date: | 2012-08-13 | | Release date: | 2013-01-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.557 Å) | | Cite: | Structural Basis for dsRNA Recognition, Filament Formation, and Antiviral Signal Activation by MDA5.
Cell(Cambridge,Mass.), 152, 2013
|
|
7TDX
 
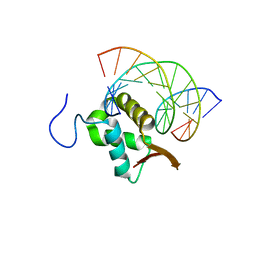 | | Structure of FOXP3-DNA complex | | Descriptor: | DNA (5'-D(*AP*AP*AP*TP*TP*TP*GP*TP*TP*TP*AP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*TP*AP*AP*AP*CP*AP*AP*AP*TP*TP*T)-3'), Forkhead box P3 | | Authors: | Leng, F, Hur, S. | | Deposit date: | 2022-01-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The transcription factor FoxP3 can fold into two dimerization states with divergent implications for regulatory T cell function and immune homeostasis.
Immunity, 55, 2022
|
|
7TDW
 
 | | Structure of FOXP3-DNA complex | | Descriptor: | DNA (5'-D(*AP*AP*AP*TP*TP*TP*GP*TP*TP*TP*AP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*TP*AP*AP*AP*CP*AP*AP*AP*TP*TP*T)-3'), Forkhead box P3 | | Authors: | Leng, F, Hur, S. | | Deposit date: | 2022-01-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The transcription factor FoxP3 can fold into two dimerization states with divergent implications for regulatory T cell function and immune homeostasis.
Immunity, 55, 2022
|
|
3J6J
 
 | | 3.6 Angstrom resolution MAVS filament generated from helical reconstruction | | Descriptor: | Mitochondrial antiviral-signaling protein | | Authors: | Wu, B, Peisley, A, Li, Z, Egelman, E, Walz, T, Penczek, P, Hur, S. | | Deposit date: | 2014-03-13 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Molecular Imprinting as a Signal-Activation Mechanism of the Viral RNA Sensor RIG-I.
Mol.Cell, 55, 2014
|
|
