7N8R
 
 | | FGTGFG segment from the Nucleoporin p54, residues 63-68 | | 分子名称: | FGTGFG segment from the Nucleoporin p54, residues 63-68, trifluoroacetic acid | | 著者 | Hughes, M.P, Sawaya, M.R, Eisenberg, D.S. | | 登録日 | 2021-06-15 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Extended beta-Strands Contribute to Reversible Amyloid Formation.
Acs Nano, 16, 2022
|
|
8V1L
 
 | |
8TH5
 
 | |
8TH1
 
 | |
8TH6
 
 | | Crystal Structure of the G3BP1 NTF2-like domain bound to USP10 peptide | | 分子名称: | 1,2-ETHANEDIOL, Ras GTPase-activating protein-binding protein 1, Ubiquitin carboxyl-terminal hydrolase 10 | | 著者 | Hughes, M.P, Taylor, J.P, Yang, Z. | | 登録日 | 2023-07-14 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Interaction between host G3BP and viral nucleocapsid protein regulates SARS-CoV-2 replication and pathogenicity.
Cell Rep, 43, 2024
|
|
6BZP
 
 | | STGGYG from low-complexity domain of FUS, residues 77-82 | | 分子名称: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, RNA-binding protein FUS | | 著者 | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Gonen, T, Eisenberg, D.S. | | 登録日 | 2017-12-25 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (1.1 Å) | | 主引用文献 | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
6BZM
 
 | | GFGNFGTS from low-complexity/FG repeat domain of Nup98, residues 116-123 | | 分子名称: | Nuclear pore complex protein Nup98-Nup96 | | 著者 | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Chong, L, Gonen, T, Eisenberg, D.S. | | 登録日 | 2017-12-24 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (0.9 Å) | | 主引用文献 | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
1LO2
 
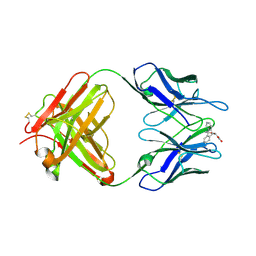 | | Retro-Diels-Alderase Catalytic Antibody | | 分子名称: | If kappa light chain, Ig gamma 2a heavy chain, [2'-CARBOXYLETHYL]-10-METHYL-ANTHRACENE ENDOPEROXIDE | | 著者 | Hugot, M, Reymond, J.L, Baumann, U. | | 登録日 | 2002-05-06 | | 公開日 | 2002-06-26 | | 最終更新日 | 2021-07-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A structural basis for the activity of retro-Diels-Alder catalytic antibodies: evidence for a catalytic aromatic residue.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LO4
 
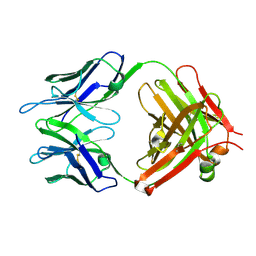 | |
1LO0
 
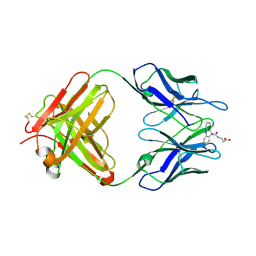 | | Catalytic Retro-Diels-Alderase Transition State Analogue Complex | | 分子名称: | 3-{[(9-CYANO-9,10-DIHYDRO-10-METHYLACRIDIN-9-YL)CARBONYL]AMINO}PROPANOIC ACID, If kappa light chain, Ig gamma 2a heavy chain | | 著者 | Hugot, M, Reymond, J.L, Baumann, U. | | 登録日 | 2002-05-05 | | 公開日 | 2002-06-26 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A structural basis for the activity of retro-Diels-Alder catalytic antibodies: evidence for a catalytic aromatic residue.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6BWZ
 
 | | SYSGYS from low-complexity domain of FUS, residues 37-42 | | 分子名称: | SYSGYS peptide from low-complexity domain of FUS | | 著者 | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Tamir, G, Eisenberg, D.S. | | 登録日 | 2017-12-15 | | 公開日 | 2018-04-04 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
6BXV
 
 | | SYSSYGQS from low-complexity domain of FUS, residues 54-61 | | 分子名称: | FUS | | 著者 | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Gonen, T, Eisenberg, D.S. | | 登録日 | 2017-12-19 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
6BXX
 
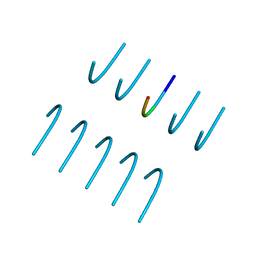 | | GYNGFG from low-complexity domain of hnRNPA1, residues 243-248 | | 分子名称: | hnRNPA1 | | 著者 | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Gonen, T, Eisenberg, D.S. | | 登録日 | 2017-12-19 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
1LO3
 
 | |
8TH7
 
 | |
6T13
 
 | | CRYSTAL STRUCTURE OF GLUCOCEREBROSIDASE IN COMPLEX WITH A PYRROLOPYRAZINE | | 分子名称: | 1,2-ETHANEDIOL, 1-[4-[2-(4-methoxyphenyl)-5-methyl-pyrrolo[2,3-b]pyrazin-6-yl]piperidin-1-yl]ethanone, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Benz, J, Ehler, A, Hug, M, Huber, S, Rufer, A.C, Guba, W, Jagasia, R, Hofmann, E.C, Rodriguez Sarmiento, R.M. | | 登録日 | 2019-10-03 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Novel beta-Glucocerebrosidase Activators That Bind to a New Pocket at a Dimer Interface and Induce Dimerization.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6WQK
 
 | | hnRNPA2 Low complexity domain (LCD) determined by cryoEM | | 分子名称: | MCherry fluorescent protein,Heterogeneous nuclear ribonucleoproteins A2/B1 chimera | | 著者 | Lu, J, Cao, Q, Hughes, M.P, Sawaya, M.R, Boyer, D.R, Cascio, D, Eisenberg, D.S. | | 登録日 | 2020-04-29 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | CryoEM structure of the low-complexity domain of hnRNPA2 and its conversion to pathogenic amyloid.
Nat Commun, 11, 2020
|
|
6WPQ
 
 | | GNYNVF from hnRNPA2-low complexity domain segment, residues 286-291, D290V variant | | 分子名称: | Heterogeneous nuclear ribonucleoprotein A2 | | 著者 | Lu, J, Cao, Q, Hughes, M.P, Sawaya, M.R, Boyer, D.R, Cascio, D, Eisenberg, D.S. | | 登録日 | 2020-04-27 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | CryoEM structure of the low-complexity domain of hnRNPA2 and its conversion to pathogenic amyloid.
Nat Commun, 11, 2020
|
|
6ST5
 
 | | crystal structure of LicM2 | | 分子名称: | GLYCEROL, LicM2, MAGNESIUM ION, ... | | 著者 | Gonsior, M, Mainz, A, Hugelland, M, Kuthning, A, Tietzmann, M, Dobbek, H, Martins, B.M, Sussmuth, R. | | 登録日 | 2019-09-10 | | 公開日 | 2022-08-10 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | crystal structure of LicM2
To Be Published
|
|
5MY3
 
 | | Crystal structure of the RhoGAP domain of Rgd1p at 2.19 Angstroms resolution | | 分子名称: | RHO GTPase-activating protein RGD1 | | 著者 | Martinez, D.M, d'Estaintot, B.L, Granier, T, Hugues, M, Odaert, B, Gallois, B, Doignon, F. | | 登録日 | 2017-01-25 | | 公開日 | 2018-01-24 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Structural evidence of a phosphoinositide-binding site in the Rgd1-RhoGAP domain.
Biochem. J., 474, 2017
|
|
1CBG
 
 | | THE CRYSTAL STRUCTURE OF A CYANOGENIC BETA-GLUCOSIDASE FROM WHITE CLOVER (TRIFOLIUM REPENS L.), A FAMILY 1 GLYCOSYL-HYDROLASE | | 分子名称: | CYANOGENIC BETA-GLUCOSIDASE | | 著者 | Barrett, T.E, Suresh, C.G, Tolley, S.P, Hughes, M.A. | | 登録日 | 1995-07-31 | | 公開日 | 1995-10-15 | | 最終更新日 | 2019-08-14 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The crystal structure of a cyanogenic beta-glucosidase from white clover, a family 1 glycosyl hydrolase.
Structure, 3, 1995
|
|
