1HO4
 
 | | CRYSTAL STRUCTURE OF PYRIDOXINE 5'-PHOSPHATE SYNTHASE IN COMPLEX WITH PYRIDOXINE 5'-PHOSPHATE AND INORGANIC PHOSPHATE | | Descriptor: | PHOSPHATE ION, PYRIDOXINE 5'-PHOSPHATE SYNTHASE, PYRIDOXINE-5'-PHOSPHATE | | Authors: | Garrido-Franco, M, Laber, B, Huber, R, Clausen, T. | | Deposit date: | 2000-12-08 | | Release date: | 2001-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the function of pyridoxine 5'-phosphate synthase.
Structure, 9, 2001
|
|
1R2K
 
 | | Crystal structure of MoaB from Escherichia coli | | Descriptor: | Molybdenum cofactor biosynthesis protein B, SULFATE ION | | Authors: | Bader, G, Gomez-Ortiz, M, Haussmann, C, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2003-09-28 | | Release date: | 2004-06-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the molybdenum-cofactor biosynthesis protein MoaB of Escherichia coli.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1FS8
 
 | | CYTOCHROME C NITRITE REDUCTASE FROM WOLINELLA SUCCINOGENES-SULFATE COMPLEX | | Descriptor: | ACETATE ION, CALCIUM ION, CYTOCHROME C NITRITE REDUCTASE, ... | | Authors: | Einsle, O, Stach, P, Messerschmidt, A, Simon, J, Kroeger, A, Huber, R, Kroneck, P.M.H. | | Deposit date: | 2000-09-08 | | Release date: | 2001-01-17 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cytochrome c nitrite reductase from Wolinella succinogenes. Structure at 1.6 A resolution, inhibitor binding, and heme-packing motifs.
J.Biol.Chem., 275, 2000
|
|
1FS9
 
 | | CYTOCHROME C NITRITE REDUCTASE FROM WOLINELLA SUCCINOGENES-AZIDE COMPLEX | | Descriptor: | AZIDE ION, CALCIUM ION, CYTOCHROME C NITRITE REDUCTASE, ... | | Authors: | Einsle, O, Stach, P, Messerschmidt, A, Simon, J, Kroeger, A, Huber, R, Kroneck, P.M.H. | | Deposit date: | 2000-09-08 | | Release date: | 2001-01-17 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cytochrome c nitrite reductase from Wolinella succinogenes. Structure at 1.6 A resolution, inhibitor binding, and heme-packing motifs.
J.Biol.Chem., 275, 2000
|
|
1FS7
 
 | | CYTOCHROME C NITRITE REDUCTASE FROM WOLINELLA SUCCINOGENES | | Descriptor: | ACETATE ION, CALCIUM ION, CYTOCHROME C NITRITE REDUCTASE, ... | | Authors: | Einsle, O, Stach, P, Messerschmidt, A, Simon, J, Kroeger, A, Huber, R, Kroneck, P.M.H. | | Deposit date: | 2000-09-08 | | Release date: | 2001-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cytochrome c nitrite reductase from Wolinella succinogenes. Structure at 1.6 A resolution, inhibitor binding, and heme-packing motifs.
J.Biol.Chem., 275, 2000
|
|
1FFU
 
 | | CARBON MONOXIDE DEHYDROGENASE FROM HYDROGENOPHAGA PSEUDOFLAVA WHICH LACKS THE MO-PYRANOPTERIN MOIETY OF THE MOLYBDENUM COFACTOR | | Descriptor: | CUTL, MOLYBDOPROTEIN OF CARBON MONOXIDE DEHYDROGENASE, CUTM, ... | | Authors: | Haenzelmann, P, Dobbek, H, Gremer, L, Huber, R, Meyer, O. | | Deposit date: | 2000-07-26 | | Release date: | 2000-09-15 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The effect of intracellular molybdenum in Hydrogenophaga pseudoflava on the crystallographic structure of the seleno-molybdo-iron-sulfur flavoenzyme carbon monoxide dehydrogenase.
J.Mol.Biol., 301, 2000
|
|
1PRC
 
 | | CRYSTALLOGRAPHIC REFINEMENT AT 2.3 ANGSTROMS RESOLUTION AND REFINED MODEL OF THE PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS | | Descriptor: | 15-trans-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Deisenhofer, J, Epp, O, Miki, K, Huber, R, Michel, H. | | Deposit date: | 1988-02-04 | | Release date: | 1989-01-09 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic refinement at 2.3 A resolution and refined model of the photosynthetic reaction centre from Rhodopseudomonas viridis.
J.Mol.Biol., 246, 1995
|
|
1G71
 
 | | CRYSTAL STRUCTURE OF PYROCOCCUS FURIOSUS DNA PRIMASE | | Descriptor: | CHLORIDE ION, DNA PRIMASE, SULFATE ION, ... | | Authors: | Augustin, M.A, Huber, R, Kaiser, J.T. | | Deposit date: | 2000-11-08 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a DNA-dependent RNA polymerase (DNA primase).
Nat.Struct.Biol., 8, 2001
|
|
1ICQ
 
 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO COMPLEXED WITH 9R,13R-OPDA | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, 9R,13R-12-OXOPHYTODIENOIC ACID, FLAVIN MONONUCLEOTIDE | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
1J2P
 
 | | alpha-ring from the proteasome from archaeoglobus fulgidus | | Descriptor: | Proteasome alpha subunit | | Authors: | Groll, M, Brandstetter, H, Bartunik, H, Bourenkow, G, Huber, R. | | Deposit date: | 2003-01-08 | | Release date: | 2003-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Investigations on the Maturation and Regulation of Archaebacterial Proteasomes
J.MOL.BIOL., 327, 2003
|
|
1OBW
 
 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE | | Descriptor: | INORGANIC PYROPHOSPHATASE, MAGNESIUM ION | | Authors: | Oganessyan, V.Yu, Harutyunyan, E.H, Avaeva, S.M, Oganessyan, N.N, Mather, T, Huber, R. | | Deposit date: | 1996-10-09 | | Release date: | 1997-09-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of holo inorganic pyrophosphatase from Escherichia coli at 1.9 A resolution. Mechanism of hydrolysis.
Biochemistry, 36, 1997
|
|
1ITZ
 
 | | Maize Transketolase in complex with TPP | | Descriptor: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, Transketolase | | Authors: | Gerhardt, S, Echt, S, Bader, G, Freigang, J, Busch, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-02-15 | | Release date: | 2003-02-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and properties of an engineered transketolase from maize
PLANT PHYSIOL., 132, 2003
|
|
1J93
 
 | | Crystal Structure and Substrate Binding Modeling of the Uroporphyrinogen-III Decarboxylase from Nicotiana tabacum: Implications for the Catalytic Mechanism | | Descriptor: | SULFATE ION, UROPORPHYRINOGEN DECARBOXYLASE | | Authors: | Martins, B.M, Grimm, B, Mock, H.-P, Huber, R, Messerschmidt, A. | | Deposit date: | 2001-05-23 | | Release date: | 2001-10-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and substrate binding modeling of the uroporphyrinogen-III decarboxylase from Nicotiana tabacum. Implications for the catalytic mechanism
J.Biol.Chem., 276, 2001
|
|
1JG5
 
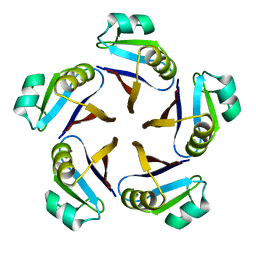 | | CRYSTAL STRUCTURE OF RAT GTP CYCLOHYDROLASE I FEEDBACK REGULATORY PROTEIN, GFRP | | Descriptor: | GTP CYCLOHYDROLASE I FEEDBACK REGULATORY PROTEIN, POTASSIUM ION | | Authors: | Bader, G, Schiffmann, S, Herrmann, A, Fischer, M, Gutlich, M, Auerbach, G, Ploom, T, Bacher, A, Huber, R, Lemm, T. | | Deposit date: | 2001-06-23 | | Release date: | 2001-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of rat GTP cyclohydrolase I feedback regulatory protein, GFRP.
J.Mol.Biol., 312, 2001
|
|
1JHJ
 
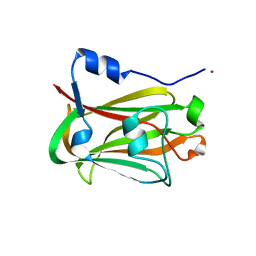 | | Crystal structure of the APC10/Doc1 subunit of the human anaphase-promoting complex | | Descriptor: | APC10, NICKEL (II) ION | | Authors: | Wendt, K.S, Vodermaier, H.C, Jacob, U, Gieffers, C, Gmachl, M, Peters, J.-M, Huber, R, Sondermann, P. | | Deposit date: | 2001-06-28 | | Release date: | 2001-10-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the APC10/DOC1 subunit of the human anaphase-promoting complex
Nat.Struct.Biol., 8, 2001
|
|
1JQG
 
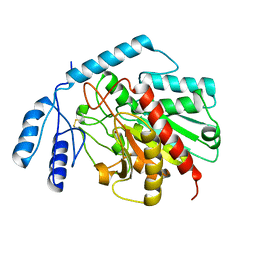 | | Crystal Structure of the Carboxypeptidase A from Helicoverpa Armigera | | Descriptor: | ZINC ION, carboxypeptidase A | | Authors: | Estebanez-Perpina, E, Bayes, A, Vendrell, J, Jongsma, M.A, Bown, D.P, Gatehouse, J.A, Huber, R, Bode, W, Aviles, F.X, Reverter, D. | | Deposit date: | 2001-08-07 | | Release date: | 2002-08-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a novel mid-gut procarboxypeptidase from the cotton pest Helicoverpa armigera.
J.Mol.Biol., 313, 2001
|
|
1RFN
 
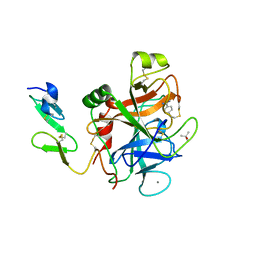 | | HUMAN COAGULATION FACTOR IXA IN COMPLEX WITH P-AMINO BENZAMIDINE | | Descriptor: | CALCIUM ION, P-AMINO BENZAMIDINE, PROTEIN (COAGULATION FACTOR IX), ... | | Authors: | Hopfner, K.-P, Lang, A, Karcher, A, Sichler, K, Kopetzki, E, Brandstetter, H, Huber, R, Bode, W, Engh, R.A. | | Deposit date: | 1999-04-19 | | Release date: | 1999-09-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Coagulation factor IXa: the relaxed conformation of Tyr99 blocks substrate binding.
Structure Fold.Des., 7, 1999
|
|
1HH2
 
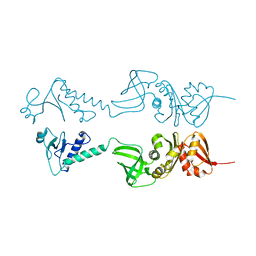 | | Crystal structure of NusA from Thermotoga maritima | | Descriptor: | N UTILIZATION SUBSTANCE PROTEIN A | | Authors: | Worbs, M, Bourenkov, G.P, Bartunik, H.D, Huber, R, Wahl, M.C. | | Deposit date: | 2000-12-18 | | Release date: | 2001-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An Extended RNA Binding Surface Through Arrayed S1 and Kh Domains in Transcription Factor Nusa
Mol.Cell, 7, 2001
|
|
1GTQ
 
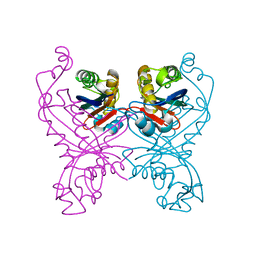 | | 6-PYRUVOYL TETRAHYDROPTERIN SYNTHASE | | Descriptor: | 6-PYRUVOYL TETRAHYDROPTERIN SYNTHASE, ZINC ION | | Authors: | Nar, H, Huber, R, Heizmann, C.W, Thoeny, B, Buergisser, D. | | Deposit date: | 1995-09-16 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of 6-pyruvoyl tetrahydropterin synthase, an enzyme involved in tetrahydrobiopterin biosynthesis.
EMBO J., 13, 1994
|
|
2H6T
 
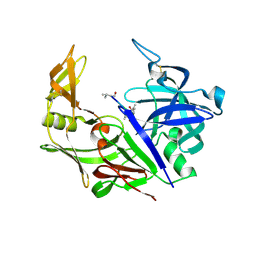 | | Secreted aspartic proteinase (Sap) 3 from Candida albicans complexed with pepstatin A | | Descriptor: | Candidapepsin-3, ZINC ION, pepstatin A | | Authors: | Ruge, E, Borelli, C, Maskos, K, Huber, R. | | Deposit date: | 2006-06-01 | | Release date: | 2007-06-12 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the secreted aspartic proteinase 3 from Candida albicans and its complex with pepstatin A.
Proteins, 68, 2007
|
|
2H6S
 
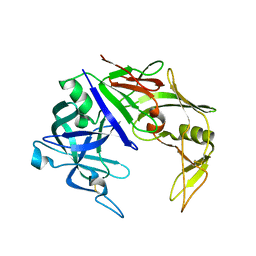 | | Secreted aspartic proteinase (Sap) 3 from Candida albicans | | Descriptor: | Candidapepsin-3, ZINC ION | | Authors: | Ruge, E, Borelli, C, Maskos, K, Huber, R. | | Deposit date: | 2006-06-01 | | Release date: | 2007-06-12 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the secreted aspartic proteinase 3 from Candida albicans and its complex with pepstatin A.
Proteins, 68, 2007
|
|
1RYP
 
 | | CRYSTAL STRUCTURE OF THE 20S PROTEASOME FROM YEAST AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | 20S PROTEASOME, MAGNESIUM ION | | Authors: | Groll, M, Ditzel, L, Loewe, J, Stock, D, Bochtler, M, Bartunik, H.D, Huber, R. | | Deposit date: | 1997-02-26 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of 20S proteasome from yeast at 2.4 A resolution.
Nature, 386, 1997
|
|
2F9O
 
 | | Crystal Structure of the Recombinant Human Alpha I Tryptase Mutant D216G | | Descriptor: | Tryptase alpha-1, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Rohr, K.B, Selwood, T, Marquardt, U, Huber, R, Schechter, N.M, Bode, W, Than, M.E. | | Deposit date: | 2005-12-06 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structures of Free and Leupeptin-complexed Human alpha I-Tryptase Mutants: Indication for an alpha to beta-Tryptase Transition
J.Mol.Biol., 357, 2005
|
|
1P0K
 
 | | IPP:DMAPP isomerase type II apo structure | | Descriptor: | Isopentenyl-diphosphate delta-isomerase, PHOSPHATE ION | | Authors: | Steinbacher, S, Kaiser, J, Gerhardt, S, Eisenreich, W, Huber, R, Bacher, A, Rohdich, F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-06-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Type II Isopentenyl Diphosphate:Dimethylallyl Diphosphate Isomerase from Bacillus subtilis
J.Mol.Biol., 329, 2003
|
|
1ONO
 
 | | IspC Mn2+ complex | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, MANGANESE (II) ION | | Authors: | Steinbacher, S, Kaiser, J, Eisenreich, W, Huber, R, Bacher, A, Rohdich, F. | | Deposit date: | 2003-02-28 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of fosmidomycin action revealed by the complex with 2-C-methyl-D-erythritol
4-phosphate synthase (IspC). Implications for the catalytic mechanism and anti-malaria
drug development.
J.BIOL.CHEM., 278, 2003
|
|
