8IZA
 
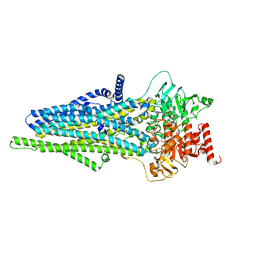 | | cryo-EM structure of ATP-bound hMRP4 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 4, MAGNESIUM ION | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-04-06 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | cryo-EM structure of ATP-bound hMRP4
To Be Published
|
|
8IZ7
 
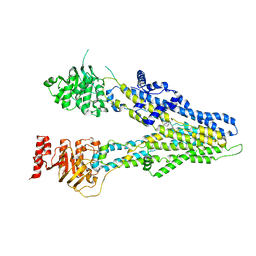 | | cryo-EM structure of sulindac-bound hMRP4 | | Descriptor: | ATP-binding cassette sub-family C member 4, [(1Z)-5-fluoro-2-methyl-1-{4-[methylsulfinyl]benzylidene}-1H-inden-3-yl]acetic acid | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-04-06 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | cryo-EM structure of sulindac-bound hMRP4
To Be Published
|
|
8IZ8
 
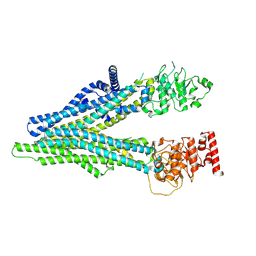 | | cryo EM structure of apo hMRP4 | | Descriptor: | ATP-binding cassette sub-family C member 4 | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-04-06 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | cryo EM structure of apo hMRP4
To Be Published
|
|
8IZ9
 
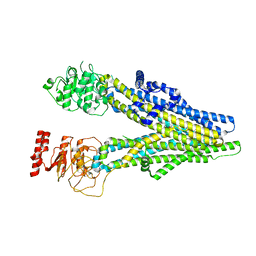 | | cryo-EM structure of PGE1-bound hMRP4 | | Descriptor: | 7-[(1R,3R)-3-hydroxy-2-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-5-oxocyclopentyl]heptanoic acid, ATP-binding cassette sub-family C member 4 | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-04-06 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | cryo-EM structure of PGE1-bound hMRP4
To Be Published
|
|
7CYQ
 
 | | Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Yan, L, Ge, J, Zheng, L, Zhang, Y, Gao, Y, Wang, T, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis.
Cell, 184, 2021
|
|
5X5Y
 
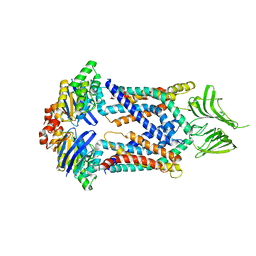 | | A membrane protein complex | | Descriptor: | Probable ATP-binding component of ABC transporter, Uncharacterized protein | | Authors: | Luo, Q, Yang, X, Huang, Y. | | Deposit date: | 2017-02-18 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.465 Å) | | Cite: | Structural basis for lipopolysaccharide extraction by ABC transporter LptB2FG
Nat. Struct. Mol. Biol., 24, 2017
|
|
5XYW
 
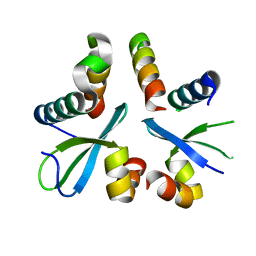 | |
5XYV
 
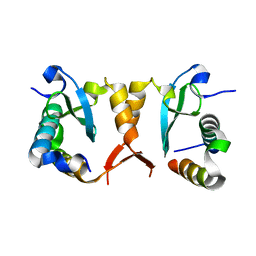 | |
5Z9X
 
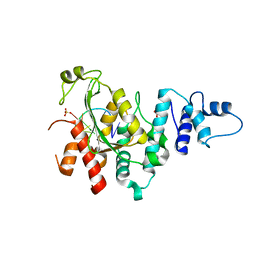 | | Arabidopsis SMALL RNA DEGRADING NUCLEASE 1 in complex with an RNA substrate | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*GP*CP*CP*CP*AP*UP*UP*AP*G)-3'), SULFATE ION, ... | | Authors: | Chen, J, Liu, L, You, C, Gu, J, Ruan, W, Zhang, L, Gan, J, Cao, C, Huang, Y, Chen, X, Ma, J. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical insights into small RNA 3' end trimming by Arabidopsis SDN1.
Nat Commun, 9, 2018
|
|
5Z9Z
 
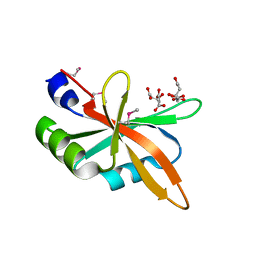 | | The C-terminal RRM domain of Arabidopsis SMALL RNA DEGRADING NUCLEASE 1 (E329A/E330A/E332A) | | Descriptor: | CITRATE ANION, Small RNA degrading nuclease 1 | | Authors: | Chen, J, Liu, L, You, C, Gu, J, Ruan, W, Zhang, L, Cao, C, Gan, J, Huang, Y, Chen, X, Ma, J. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Structural and biochemical insights into small RNA 3' end trimming by Arabidopsis SDN1.
Nat Commun, 9, 2018
|
|
5ZUM
 
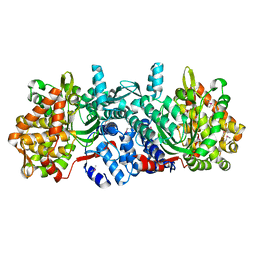 | | Structure of dipeptidyl-peptidase III from Corallococcus sp. strain EGB | | Descriptor: | ZINC ION, dipeptidyl-peptidase III | | Authors: | Zhang, H, Duan, Y.J, Li, Z.K, Liu, W.D, Huang, Y, Cui, Z.L. | | Deposit date: | 2018-05-08 | | Release date: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of dipeptidyl peptidase III from Corallococcus sp. strain EGB
To Be Published
|
|
6A6O
 
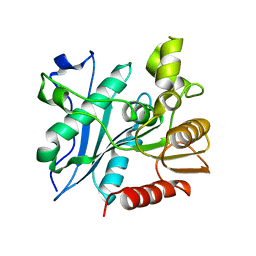 | | Crystal structure of acetyl ester-xyloside bifunctional hydrolase from Caldicellulosiruptor lactoaceticus | | Descriptor: | Esterase/lipase-like protein | | Authors: | Cao, H, Huang, Y, Sun, L.C, Liu, X, Liu, T.F, Wang, F.Z, Xin, F.J. | | Deposit date: | 2018-06-28 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the Dual-Substrate Recognition and Catalytic Mechanisms of a Bifunctional Acetyl Ester-Xyloside Hydrolase from Caldicellulosiruptor lactoaceticus.
Acs Catalysis, 9, 2019
|
|
6AIN
 
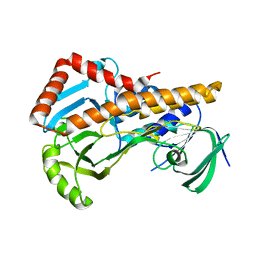 | | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PnpA | | Authors: | Chen, Q.Z, Huang, Y, Duan, Y.J, Li, Z.K, Liu, W.D, Cui, Z.L. | | Deposit date: | 2018-08-24 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4: The key enzyme involved in p-nitrophenol degradation.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
6AIO
 
 | | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4 | | Descriptor: | PnpA | | Authors: | Chen, Q.Z, Huang, Y, Duan, Y.J, Li, Z.K, Liu, W.D, Cui, Z.L. | | Deposit date: | 2018-08-24 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4: The key enzyme involved in p-nitrophenol degradation.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
7CXN
 
 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
7CXM
 
 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
7WZ6
 
 | | Crystal structure of MyoD-E47 | | Descriptor: | Isoform E47 of Transcription factor E2-alpha, Myoblast determination protein 1 | | Authors: | Zhong, J, Huang, Y, Ma, J. | | Deposit date: | 2022-02-17 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of the bHLH domains of MyoD-E47 heterodimer.
Biochem.Biophys.Res.Commun., 621, 2022
|
|
6KCU
 
 | | Shuguo PWWP domain | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LD23804p | | Authors: | Liu, Y.C, Huang, Y. | | Deposit date: | 2019-06-29 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Shuguo PWWP domain
To Be Published
|
|
6KCO
 
 | | Shuguo PWWP in complex with ssDNA | | Descriptor: | DNA (5'-D(*TP*CP*CP*CP*T)-3'), LD23804p | | Authors: | Liu, Y.C, Huang, Y. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Shuguo PWWP in complex with ssDNA
To Be Published
|
|
6KCP
 
 | | Shuguo PWWP in complex with ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*T)-3'), LD23804p | | Authors: | Liu, Y.C, Huang, Y. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Shuguo PWWP in complex with ssDNA
To Be Published
|
|
6KD6
 
 | | Shuguo PWWP domain | | Descriptor: | LD23804p | | Authors: | Liu, Y.C, Huang, Y. | | Deposit date: | 2019-07-01 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Shuguo PWWP domain
To Be Published
|
|
7JT0
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with MAC5576 | | Descriptor: | 3C-like proteinase, PHOSPHATE ION, thiophene-2-carbaldehyde | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Huang, Y, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-16 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
7JT7
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with compound 4 | | Descriptor: | 3C-like proteinase, ethyl (4R)-4-[[(2S)-4-methyl-2-[[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-2-(phenylmethoxycarbonylamino)butanoyl]amino]pentanoyl]amino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Huang, Y, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-17 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
7JST
 
 | | Crystal structure of SARS-CoV-2 3CL in apo form | | Descriptor: | 3C-like proteinase, PHOSPHATE ION | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Huang, Y, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-16 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
7Y1B
 
 | | 3.2 angstrom cryo-EM structure of extracellular region of mouse Basigin-2 in complex with the Fab fragment of antibody 6E7F1 | | Descriptor: | Heavy chain of 6E7F1, Isoform 2 of Basigin, Light chain of 6E7F1 | | Authors: | Zhang, H, Yang, X, Xue, Y, Huang, Y, Mo, X, Zhang, H, Li, N, Gao, N, Li, X, Wang, S, Gao, Y, Liao, J. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Allosteric modulation of monocarboxylate transporters 1 and 4 by targeting their chaperon Basigin
To Be Published
|
|
