6GOV
 
 | | Structure of THE RNA POLYMERASE LAMBDA-BASED ANTITERMINATION COMPLEX | | Descriptor: | 30S ribosomal protein S10, Antitermination protein N, DNA (I), ... | | Authors: | Loll, B, Krupp, F, Said, N, Huang, Y, Buerger, J, Mielke, T, Spahn, C.M.T, Wahl, M.C. | | Deposit date: | 2018-06-04 | | Release date: | 2019-02-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis for the Action of an All-Purpose Transcription Anti-termination Factor.
Mol.Cell, 74, 2019
|
|
1MIT
 
 | | RECOMBINANT CUCURBITA MAXIMA TRYPSIN INHIBITOR V (RCMTI-V) (NMR, MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | TRYPSIN INHIBITOR V | | Authors: | Cai, M, Gong, Y, Huang, Y, Liu, J, Prakash, O, Wen, L, Wen, J.J, Huang, J.-K, Krishnamoorthi, R. | | Deposit date: | 1995-10-26 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of recombinant Cucurbita maxima trypsin inhibitor-V determined by NMR spectroscopy.
Biochemistry, 35, 1996
|
|
1B3R
 
 | | RAT LIVER S-ADENOSYLHOMOCYSTEIN HYDROLASE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (S-ADENOSYLHOMOCYSTEINE HYDROLASE) | | Authors: | Hu, Y, Komoto, J, Huang, Y, Takusagawa, F, Gomi, T, Ogawa, H, Takata, Y, Fujioka, M. | | Deposit date: | 1998-12-14 | | Release date: | 1998-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of S-adenosylhomocysteine hydrolase from rat liver.
Biochemistry, 38, 1999
|
|
2V0X
 
 | | The dimerization domain of LAP2alpha | | Descriptor: | LAMINA-ASSOCIATED POLYPEPTIDE 2 ISOFORMS ALPHA/ZETA | | Authors: | Bradley, C.M, Jones, S, Huang, Y, Suzuki, Y, Kvaratskhelia, M, Hickman, A.B, Craigie, R, Dyda, F. | | Deposit date: | 2007-05-20 | | Release date: | 2007-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Dimerization of Lap2Alpha, a Component of the Nuclear Lamina.
Structure, 15, 2007
|
|
4ZS3
 
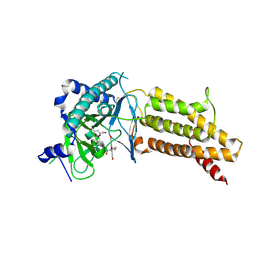 | | Structural complex of 5-aminofluorescein bound to the FTO protein | | Descriptor: | 2-OXOGLUTARIC ACID, 5-amino-2-(6-hydroxy-3-oxo-3H-xanthen-9-yl)benzoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, ... | | Authors: | Wang, T, Hong, T, Huang, Y, Yang, C.G, Zhou, X. | | Deposit date: | 2015-05-13 | | Release date: | 2015-11-04 | | Last modified: | 2015-11-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Fluorescein Derivatives as Bifunctional Molecules for the Simultaneous Inhibiting and Labeling of FTO Protein
J.Am.Chem.Soc., 137, 2015
|
|
4ZS2
 
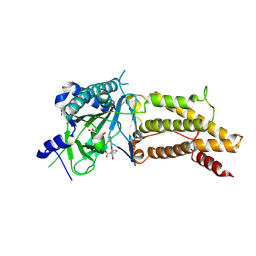 | | Structural complex of FTO/fluorescein | | Descriptor: | 2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)-BENZOIC ACID, 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase FTO, ... | | Authors: | Wang, T, Hong, T, Huang, Y, Yang, C.G, Zhou, X. | | Deposit date: | 2015-05-13 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Fluorescein Derivatives as Bifunctional Molecules for the Simultaneous Inhibiting and Labeling of FTO Protein
J.Am.Chem.Soc., 137, 2015
|
|
1OX1
 
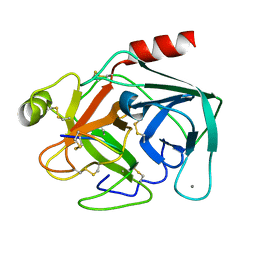 | | crystal structure of the bovine trypsin complex with a synthetic 11 peptide inhibitor | | Descriptor: | 11-mer peptide, CALCIUM ION, Trypsinogen, ... | | Authors: | Wu, G, Huang, Y, Zhu, G, Huang, Q, Tang, Y, Miyake, H, Kusunoki, M. | | Deposit date: | 2003-03-31 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure of the bovine trypsin complex with a synthetic 11 peptide inhibitor
To be published
|
|
6PUS
 
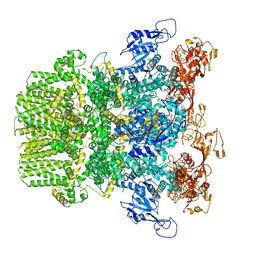 | | Human TRPM2 bound to ADPR and calcium | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CALCIUM ION, Transient receptor potential cation channel subfamily M member 2 | | Authors: | Du, J, Lu, W, Huang, Y. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ligand recognition and gating mechanism through three ligand-binding sites of human TRPM2 channel.
Elife, 8, 2019
|
|
6PUR
 
 | | Human TRPM2 bound to ADPR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Transient receptor potential cation channel subfamily M member 2 | | Authors: | Du, J, Lu, W, Huang, Y. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Ligand recognition and gating mechanism through three ligand-binding sites of human TRPM2 channel.
Elife, 8, 2019
|
|
6PUU
 
 | | Human TRPM2 bound to 8-Br-cADPR and calcium | | Descriptor: | (2R,3R,4S,5R,13R,14S,15R,16R)-24-amino-18-bromo-3,4,14,15-tetrahydroxy-7,9,11,25,26-pentaoxa-17,19,22-triaza-1-azonia-8 ,10-diphosphapentacyclo[18.3.1.1^2,5^.1^13,16^.0^17,21^]hexacosa-1(24),18,20,22-tetraene-8,10-diolate 8,10-dioxide, CALCIUM ION, Transient receptor potential cation channel subfamily M member 2 | | Authors: | Du, J, Lu, W, Huang, Y. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ligand recognition and gating mechanism through three ligand-binding sites of human TRPM2 channel.
Elife, 8, 2019
|
|
6PUO
 
 | | Human TRPM2 in the apo state | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Du, J, Lu, W, Huang, Y. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand recognition and gating mechanism through three ligand-binding sites of human TRPM2 channel.
Elife, 8, 2019
|
|
4MZ4
 
 | | Discovery of an Irreversible HCV NS5B Polymerase Inhibitor | | Descriptor: | 1-[(2-chloroquinolin-3-yl)methyl]-6-fluoro-5-methyl-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxylic acid, PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Zeng, Q, Anilkumar, G.N, Rosenblum, S.B, Huang, H.-C, Lesburg, C.A, Jiang, Y, Selyutin, O, Chan, T.-Y, Bennett, F, Chen, K.X, Venkatraman, S, Sannigrahi, M, Velazquez, F, Duca, J.S, Gavalas, S, Huang, Y, Pu, H, Wang, L, Pinto, P, Vibulbhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Li, C, Hesk, D, Gesell, J, Sorota, S, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | Deposit date: | 2013-09-29 | | Release date: | 2013-12-11 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Discovery of an irreversible HCV NS5B polymerase inhibitor.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2YOJ
 
 | | HCV NS5B polymerase complexed with pyridonylindole compound | | Descriptor: | 4-fluoranyl-6-[(7-fluoranyl-4-oxidanylidene-3H-quinazolin-6-yl)methyl]-8-(2-oxidanylidene-1H-pyridin-3-yl)furo[2,3-e]indole-7-carboxylic acid, PHOSPHATE ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Chen, K.X, Venkatraman, S, Anilkumar, G.N, Zeng, Q, Lesburg, C.A, Vibulbhan, B, Yang, W, Velazquez, F, Chan, T.-Y, Bennett, F, Sannigrahi, M, Jiang, Y, Duca, J.S, Pinto, P, Gavalas, S, Huang, Y, Wu, W, Selyutin, O, Agrawal, S, Feld, B, Huang, H.-C, Li, C, Cheng, K.-C, Shih, N.-Y, Kozlowski, J.A, Rosenblum, S.B, Njoroge, F.G. | | Deposit date: | 2012-10-24 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of Sch 900188: A Potent Hepatitis C Virus Ns5B Polymerase Inhibitor Prodrug as a Development Candidate
Acs Med.Chem.Lett., 5, 2014
|
|
6LP2
 
 | | Structure of Lpg2148/UBE2N-Ub complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 N, Uncharacterized protein lpg2148 | | Authors: | Feng, Y, Wang, Y, Huang, Y, Li, D. | | Deposit date: | 2020-01-08 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.479 Å) | | Cite: | Structure of Lpg2148/UBE2N-Ub complex
To Be Published
|
|
8YTJ
 
 | | Cryo-EM structure of enterovirus A71 mature virion | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2024-03-26 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Cryo-EM structure of enterovirus A71 mature virion in complex with Fab h1A6.2
To Be Published
|
|
8YTB
 
 | | Cryo-EM structure of enterovirus A71 empty particle | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2024-03-25 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Cryo-EM structure of enterovirus A71 empty particle
To Be Published
|
|
7BTF
 
 | | SARS-CoV-2 RNA-dependent RNA polymerase in complex with cofactors in reduced condition | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | Deposit date: | 2020-04-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
1MV4
 
 | | TM9A251-284: A Peptide Model of the C-Terminus of a Rat Striated Alpha Tropomyosin | | Descriptor: | Tropomyosin 1 alpha chain | | Authors: | Greenfield, N.J, Swapna, G.V.T, Huang, Y, Palm, T, Graboski, S, Montelione, G.T, Hitchcock-Degregori, S.E. | | Deposit date: | 2002-09-24 | | Release date: | 2003-02-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Carboxyl Terminus of Striated alpha-Tropomyosin in Solution Reveals an Unusual Parallel Arrangement of Interacting alpha-Helices
Biochemistry, 42, 2003
|
|
8IZ9
 
 | | cryo-EM structure of PGE1-bound hMRP4 | | Descriptor: | 7-[(1R,3R)-3-hydroxy-2-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-5-oxocyclopentyl]heptanoic acid, ATP-binding cassette sub-family C member 4 | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-04-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for substrate and inhibitor recognition of human multidrug transporter MRP4.
Commun Biol, 6, 2023
|
|
8IZA
 
 | | cryo-EM structure of ATP-bound hMRP4 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 4, MAGNESIUM ION | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-04-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural basis for substrate and inhibitor recognition of human multidrug transporter MRP4.
Commun Biol, 6, 2023
|
|
6LQH
 
 | | High resolution architecture of curli complex | | Descriptor: | Curli production assembly/transport component CsgF, Curli production assembly/transport component CsgG | | Authors: | Zhang, M, Shi, H, Huang, Y. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cryo-EM structure of the nonameric CsgG-CsgF complex and its implications for controlling curli biogenesis in Enterobacteriaceae.
Plos Biol., 18, 2020
|
|
6LQJ
 
 | | Low resolution architecture of curli complex | | Descriptor: | Curli production assembly/transport component CsgF, Curli production assembly/transport component CsgG | | Authors: | Zhang, M, Shi, H, Huang, Y. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Cryo-EM structure of the nonameric CsgG-CsgF complex and its implications for controlling curli biogenesis in Enterobacteriaceae.
Plos Biol., 18, 2020
|
|
4WUM
 
 | |
6M4G
 
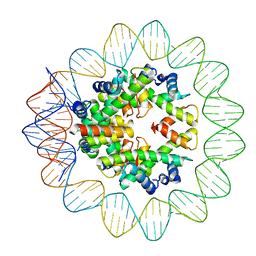 | | Structural mechanism of nucleosome dynamics governed by human histone variants H2A.B and H2A.Z.2.2 | | Descriptor: | DNA (93-MER), Histone H2A-Bbd type 2/3, Histone H2B type 2-E, ... | | Authors: | Zhou, M, Dai, L.C, Li, C.M, Shi, L.X, Huang, Y, Guo, Z.Q. | | Deposit date: | 2020-03-06 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of nucleosome dynamics modulation by histone variants H2A.B and H2A.Z.2.2.
Embo J., 40, 2021
|
|
6LYU
 
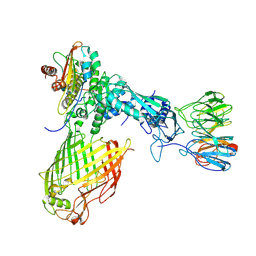 | | Structure of the BAM complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Xiao, L, Huang, Y. | | Deposit date: | 2020-02-15 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structures of the beta-barrel assembly machine recognizing outer membrane protein substrates.
Faseb J., 35, 2021
|
|
