8I3Z
 
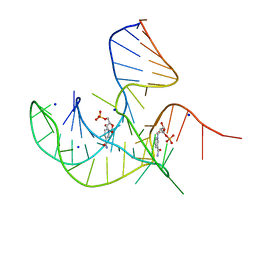 | | Crystal structure of NAD-II riboswitch (two strands) with NMN at 1.67 angstrom | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, RNA (31-MER), RNA (5'-R(*AP*GP*AP*GP*CP*GP*UP*UP*GP*CP*GP*UP*CP*CP*GP*AP*AP*AP*GP*UP*(CBV)P*GP*CP*C)-3'), ... | | Authors: | Peng, X, Lilley, D.M.J, Huang, L. | | Deposit date: | 2023-01-18 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Nucleic Acids Res., 51, 2023
|
|
5X8I
 
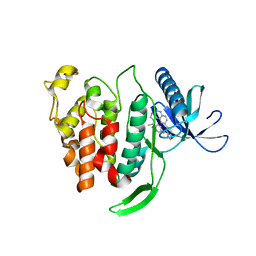 | | Crystal structure of human CLK1 in complex with compound 25 | | Descriptor: | 5-[1-[(1S)-1-(4-fluorophenyl)ethyl]-[1,2,3]triazolo[4,5-c]quinolin-8-yl]-1,3-benzoxazole, Dual specificity protein kinase CLK1 | | Authors: | Sun, Q.Z, Lin, G.F, Li, L.L, Jin, X.T, Huang, L.Y, Zhang, G, Wei, Y.Q, Lu, G.W, Yang, S.Y. | | Deposit date: | 2017-03-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Discovery of Potent and Selective Inhibitors of Cdc2-Like Kinase 1 (CLK1) as a New Class of Autophagy Inducers
J. Med. Chem., 60, 2017
|
|
3VCF
 
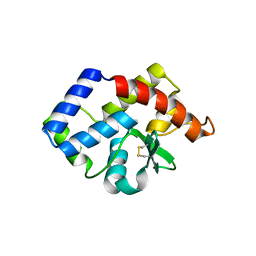 | | SSV1 integrase C-terminal catalytic domain (174-335aa) | | Descriptor: | Probable integrase | | Authors: | Ouyang, S, Liang, W, Huang, L, Liu, Z.-J. | | Deposit date: | 2012-01-04 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterization of the C-terminal catalytic domain of SSV1 integrase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6A2H
 
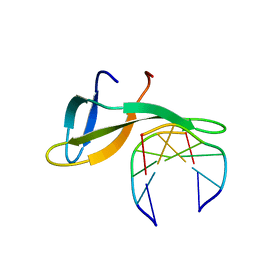 | | Architectural roles of Cren7 in folding crenarchaeal chromatin filament | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(P*AP*AP*TP*TP*AP*C)-3'), DNA (5'-D(P*GP*TP*AP*AP*TP*T)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Chen, Y.Y, Wang, L, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2018-06-11 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Architectural roles of Cren7 in folding crenarchaeal chromatin filament.
Mol. Microbiol., 111, 2019
|
|
6A2I
 
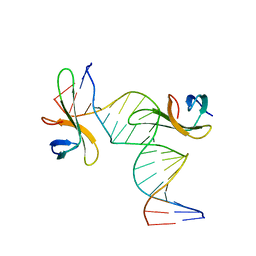 | | Architectural roles of Cren7 in folding crenarchaeal chromatin filament | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*CP*GP*TP*AP*GP*CP*TP*AP*AP*TP*TP*AP*GP*CP*TP*AP*CP*G)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Chen, Y.Y, Wang, L, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2018-06-11 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Architectural roles of Cren7 in folding crenarchaeal chromatin filament.
Mol. Microbiol., 111, 2019
|
|
6K8N
 
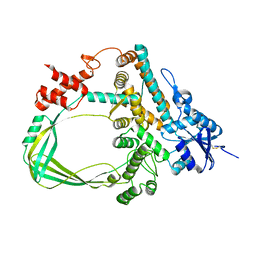 | | Crystal structure of the Sulfolobus solfataricus topoisomerase III | | Descriptor: | ZINC ION, topoisomerase III | | Authors: | Wang, H.Q, Zhang, J.H, Zheng, X, Zheng, Z.F, Dong, Y.H, Huang, L, Gong, Y. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the Sulfolobus solfataricus topoisomerase III reveal that its C-terminal novel zinc finger part is a unique decatenation domain
To Be Published
|
|
6K8O
 
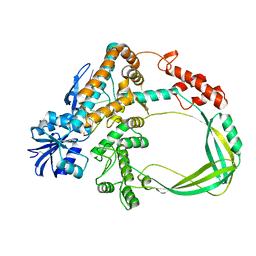 | | Crystal structure of the Sulfolobus solfataricus topoisomerase III in complex with DNA | | Descriptor: | DNA (5'-D(*GP*CP*AP*AP*GP*GP*TP*C)-3'), ZINC ION, topoisomerase III | | Authors: | Wang, H.Q, Zhang, J.H, Zheng, X, Zheng, Z.F, Dong, Y.H, Huang, L, Gong, Y. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the Sulfolobus solfataricus topoisomerase III reveal that its C-terminal novel zinc finger part is a unique decatenation domain
To Be Published
|
|
2KKE
 
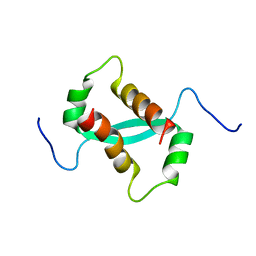 | | Solution NMR Structure of a dimeric protein of unknown function from Methanobacterium thermoautotrophicum, Northeast Structural Genomics Consortium Target TR5 | | Descriptor: | Uncharacterized protein | | Authors: | Swapna, G.V.T, Gunsalus, X, Huang, L, Xiao, K, Everett, J.K, Acton, T.B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-18 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a putative uncharacterized protein from Methanobacterium thermoautotrophicum, Northeast Structural Genomics Consortium Target:TR5
To be Published
|
|
7V9E
 
 | | Crystal structure of a methyl transferase ribozyme | | Descriptor: | BARIUM ION, GUANINE, RNA (68-MER), ... | | Authors: | Deng, J, Lilley, D.M.J, Huang, L. | | Deposit date: | 2021-08-25 | | Release date: | 2022-03-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of a methyltransferase ribozyme.
Nat.Chem.Biol., 18, 2022
|
|
7CD9
 
 | | Crystal Structure of SETDB1 tudor domain in complexed with Compound 6 | | Descriptor: | 3-methyl-2-[[(3R,5R)-1-methyl-5-(4-phenylmethoxyphenyl)piperidin-3-yl]amino]-5H-pyrrolo[3,2-d]pyrimidin-4-one, CITRIC ACID, Histone-lysine N-methyltransferase SETDB1 | | Authors: | Xiong, L, Guo, Y, Mao, X, Huang, L, Wu, C, Yang, S. | | Deposit date: | 2020-06-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7QUJ
 
 | | Structure of NsNEPS2, a 7S-cis-trans nepetalactone synthase | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NsNEPS2 | | Authors: | Hernandez Lozada, N.J, Hong, B, Wood, J.C, Caputi, L, Basquin, J, Chuang, L, Kunert, M, Rodriguez Lopez, C.R, Langley, C, Zhao, D, Buell, C.R, Lichman, B.R, O'Connor, S.E. | | Deposit date: | 2022-01-18 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biocatalytic routes to stereo-divergent iridoids.
Nat Commun, 13, 2022
|
|
3D2R
 
 | | Crystal structure of pyruvate dehydrogenase kinase isoform 4 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, L.C, Tso, S.-C, Li, J, Chuang, D.T. | | Deposit date: | 2008-05-08 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Pyruvate Dehydrogenase Kinase-4 Structures Reveal a Metastable Open Conformation Fostering Robust Core-free Basal Activity.
J.Biol.Chem., 283, 2008
|
|
2ZB4
 
 | | Crystal structure of human 15-ketoprostaglandin delta-13-reductase in complex with NADP and 15-keto-PGE2 | | Descriptor: | (5E,13E)-11-HYDROXY-9,15-DIOXOPROSTA-5,13-DIEN-1-OIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prostaglandin reductase 2, ... | | Authors: | Wu, Y.H, Wang, A.H.J, Ko, T.P, Guo, R.T, Hu, S.M, Chuang, L.M. | | Deposit date: | 2007-10-16 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2.
Structure, 16, 2008
|
|
2ZB3
 
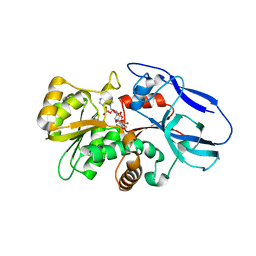 | | Crystal structure of mouse 15-ketoprostaglandin delta-13-reductase in complex with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prostaglandin reductase 2 | | Authors: | Wu, Y.H, Wang, A.H.J, Ko, T.P, Guo, R.T, Hu, S.M, Chuang, L.M. | | Deposit date: | 2007-10-16 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2.
Structure, 16, 2008
|
|
2ZB8
 
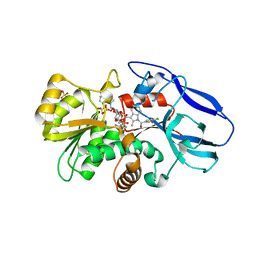 | | Crystal structure of human 15-ketoprostaglandin delta-13-reductase in complex with NADP and indomethacin | | Descriptor: | INDOMETHACIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prostaglandin reductase 2, ... | | Authors: | Wu, Y.H, Wang, A.H.J, Ko, T.P, Guo, R.T, Hu, S.M, Chuang, L.M. | | Deposit date: | 2007-10-16 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2.
Structure, 16, 2008
|
|
2ZB7
 
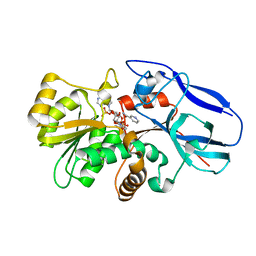 | | Crystal structure of human 15-ketoprostaglandin delta-13-reductase in complex with NADPH and nicotinamide | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE, Prostaglandin reductase 2 | | Authors: | Wu, Y.H, Wang, A.H.J, Ko, T.P, Guo, R.T, Hu, S.M, Chuang, L.M. | | Deposit date: | 2007-10-16 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2.
Structure, 16, 2008
|
|
7VT0
 
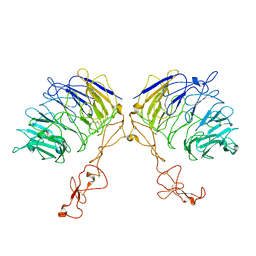 | | Dimer structure of SORLA | | Descriptor: | Sortilin-related receptor | | Authors: | Xi, Z, Cang, W, Chuang, L. | | Deposit date: | 2021-10-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal distinct apo conformations of sortilin-related receptor SORLA.
Biochem.Biophys.Res.Commun., 600, 2022
|
|
