4WBE
 
 | |
5YQ7
 
 | | Cryo-EM structure of the RC-LH core complex from Roseiflexus castenholzii | | Descriptor: | 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,14,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, BACTERIOCHLOROPHYLL A, ... | | Authors: | Shi, Y, Xin, Y.Y, Niu, T.X, Wang, Q.Q, Niu, W.Q, Huang, X.J, Ding, W, Blankenship, R.E, Xu, X.L, Sun, F. | | Deposit date: | 2017-11-05 | | Release date: | 2018-05-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the RC-LH core complex from an early branching photosynthetic prokaryote.
Nat Commun, 9, 2018
|
|
7JHI
 
 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 iodide-derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-18 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
7JHK
 
 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 in unliganded form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3436 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
7EGL
 
 | | Bicarbonate transporter complex SbtA-SbtB bound to HCO3- | | Descriptor: | BICARBONATE ION, Membrane-associated protein SbtB, SODIUM ION, ... | | Authors: | Fang, S, Huang, X, Zhang, X, Zhang, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular mechanism underlying transport and allosteric inhibition of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EJH
 
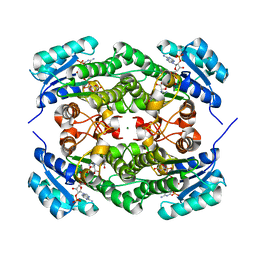 | | Crystal structure of KRED mutant-F147L/L153Q/Y190P/L199A/M205F/M206F and 2-hydroxyisoindoline-1,3-dione complex | | Descriptor: | 2-oxidanylisoindole-1,3-dione, 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase, MAGNESIUM ION, ... | | Authors: | Cui, J, Huang, X, Wang, B, Zhao, H, Zhou, J. | | Deposit date: | 2021-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72883928 Å) | | Cite: | Photoinduced chemomimetic biocatalysis for enantioselective intermolecular radical conjugate addition
Nat Catal, 2022
|
|
7EJJ
 
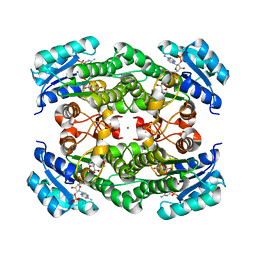 | | Crystal structure of KRED F147L/L153Q/Y190P variant and methyl methacrylate complex | | Descriptor: | 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cui, J, Huang, X, Wang, B, Zhao, H, Zhou, J. | | Deposit date: | 2021-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.80000663 Å) | | Cite: | Photoinduced chemomimetic biocatalysis for enantioselective intermolecular radical conjugate addition
Nat Catal, 2022
|
|
7F7I
 
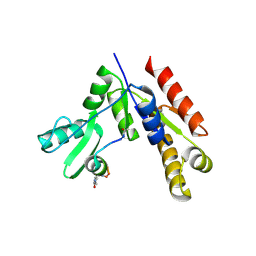 | | Stapled Peptide Inhibitor in complex with PSD95 GK domain | | Descriptor: | ACE-ARG-ILE-ARG-ARG-ASP-GLU-TYR-LEU-LYZ-ALA-ILE-GLN-NH2, Disks large homolog 4 | | Authors: | Shang, Y, Huang, X, Li, X, Zhang, M. | | Deposit date: | 2021-06-29 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Entropy of stapled peptide inhibitors in free state is the major contributor to the improvement of binding affinity with the GK domain.
Rsc Chem Biol, 2, 2021
|
|
7VE7
 
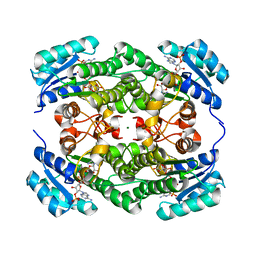 | | Crystal structure of KRED mutant-F147L/L153Q/Y190P/L199A/M205F/M206F | | Descriptor: | 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cui, J, Huang, X, Wang, B, Zhao, H, Zhou, J. | | Deposit date: | 2021-09-08 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.720007 Å) | | Cite: | Photoinduced chemomimetic biocatalysis for enantioselective intermolecular radical conjugate addition
Nat Catal, 2022
|
|
7VDO
 
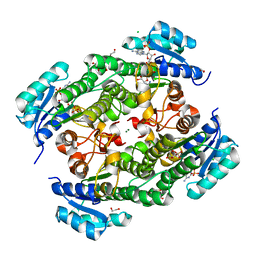 | | Crystal structure of KRED F147L/L153Q/Y190P variant | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase, ... | | Authors: | Cui, J, Huang, X, Wang, B, Zhao, H, Zhou, J. | | Deposit date: | 2021-09-07 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85571563 Å) | | Cite: | Photoinduced chemomimetic biocatalysis for enantioselective intermolecular radical conjugate addition
Nat Catal, 2022
|
|
7EJI
 
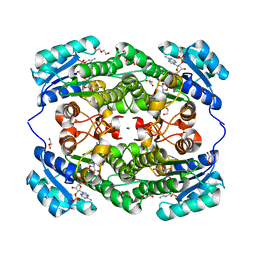 | | Crystal structure of KRED F147L/L153Q/Y190P/L199A/M205F/M206F variant and methyl methacrylate complex | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-oxidanylisoindole-1,3-dione, ... | | Authors: | Cui, J, Huang, X, Wang, B, Zhao, H, Zhou, J. | | Deposit date: | 2021-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.560016 Å) | | Cite: | Photoinduced chemomimetic biocatalysis for enantioselective intermolecular radical conjugate addition
Nat Catal, 2022
|
|
7F7G
 
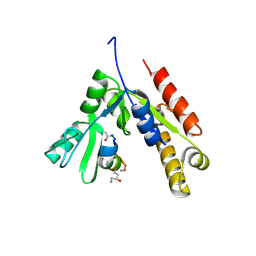 | | a linear Peptide Inhibitors in complex with GK domain | | Descriptor: | DLG4 GK domain, UNK-ARG-ILE-ARG-ARG-ASP-GLU-TYR-LEU-LYS-ALA-ILE-GLN-UNK | | Authors: | Shang, Y, Huang, X, Li, X, Zhang, M. | | Deposit date: | 2021-06-29 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.446 Å) | | Cite: | Entropy of stapled peptide inhibitors in free state is the major contributor to the improvement of binding affinity with the GK domain.
Rsc Chem Biol, 2, 2021
|
|
4WT2
 
 | | Co-crystal Structure of MDM2 in Complex with AM-7209 | | Descriptor: | 4-({[(3R,5R,6S)-1-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(4-chloro-3-fluorophenyl)-5-(3-chlorophenyl)-3-methyl-2-oxopiperidin-3-yl]acetyl}amino)-2-methoxybenzoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-10-30 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery of AM-7209, a Potent and Selective 4-Amidobenzoic Acid Inhibitor of the MDM2-p53 Interaction.
J.Med.Chem., 57, 2014
|
|
5GMJ
 
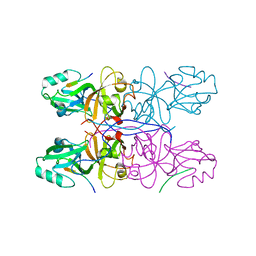 | | Crystal Structure of GRASP55 GRASP domain in complex with JAM-B C-terminus | | Descriptor: | Golgi reassembly-stacking protein 2, Junctional adhesion molecule B | | Authors: | Shi, N, Shi, X, Morelli, X, Betzi, S, Huang, X. | | Deposit date: | 2016-07-14 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.986 Å) | | Cite: | Genetic, structural, and chemical insights into the dual function of GRASP55 in germ cell Golgi remodeling and JAM-C polarized localization during spermatogenesis
PLoS Genet., 13, 2017
|
|
5GMI
 
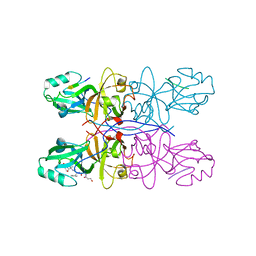 | | Crystal Structure of GRASP55 GRASP domain in complex with JAM-C C-terminus | | Descriptor: | Golgi reassembly-stacking protein 2, Junctional adhesion molecule C | | Authors: | Shi, N, Shi, X, Morelli, X, Betzi, S, Huang, X. | | Deposit date: | 2016-07-14 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Genetic, structural, and chemical insights into the dual function of GRASP55 in germ cell Golgi remodeling and JAM-C polarized localization during spermatogenesis
PLoS Genet., 13, 2017
|
|
5GML
 
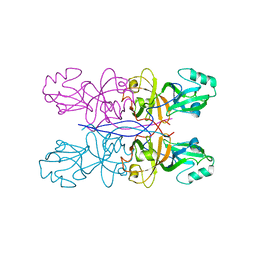 | |
5TIO
 
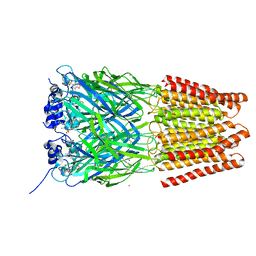 | | Crystal Structure of Human Glycine Receptor alpha-3 Bound to AM-3607 | | Descriptor: | (3S,3aS,9bS)-2-[(2H-1,3-benzodioxol-5-yl)sulfonyl]-3,5-dimethyl-1,2,3,3a,5,9b-hexahydro-4H-pyrrolo[3,4-c][1,6]naphthyridin-4-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Shaffer, P.L, Huang, X, Chen, H. | | Deposit date: | 2016-10-03 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal Structures of Human GlyRa3 Bound to a Novel Class of Potentiators with Efficacy in a Mouse Model of Neuropathic Pain
To Be Published
|
|
9AXC
 
 | | Activated CRAF/MEK heterotetramer from focused refinement of CRAF/MEK/14-3-3 complex | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, GST26/CRAF chimera, N-[3-fluoro-4-({7-[(3-fluoropyridin-2-yl)oxy]-4-methyl-2-oxo-2H-1-benzopyran-3-yl}methyl)pyridin-2-yl]-N'-methylsulfuric diamide | | Authors: | Quade, B, Cohen, S.E, Huang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | The Pan-RAF-MEK Nondegrading Molecular Glue NST-628 Is a Potent and Brain-Penetrant Inhibitor of the RAS-MAPK Pathway with Activity across Diverse RAS- and RAF-Driven Cancers.
Cancer Discov, 14, 2024
|
|
9AXA
 
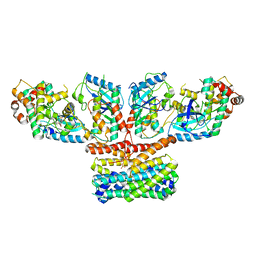 | | CryoEM structure of activated CRAF/MEK/14-3-3 complex with NST-628 | | Descriptor: | 14-3-3 protein sigma, Dual specificity mitogen-activated protein kinase kinase 1, GST26/CRAF chimera, ... | | Authors: | Quade, B, Cohen, S.E, Huang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | The Pan-RAF-MEK Nondegrading Molecular Glue NST-628 Is a Potent and Brain-Penetrant Inhibitor of the RAS-MAPK Pathway with Activity across Diverse RAS- and RAF-Driven Cancers.
Cancer Discov, 14, 2024
|
|
7WJS
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y13157 | | Descriptor: | 2-(2-cyclobutyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-07 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WKY
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y13153 | | Descriptor: | 2-(2-cyclopentyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-12 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WMU
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13146 | | Descriptor: | 1,2-ETHANEDIOL, Isoform 4 of Bromodomain-containing protein 2, ~{N}-[4-[2,4-bis(fluoranyl)phenoxy]-3-[2-(2-cyclobutyl-1~{H}-imidazol-5-yl)-5-methyl-4-oxidanylidene-furo[3,2-c]pyridin-7-yl]phenyl]ethanesulfonamide | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-17 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WN5
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13142 | | Descriptor: | 1,2-ETHANEDIOL, Isoform 4 of Bromodomain-containing protein 2, POTASSIUM ION, ... | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-17 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WNI
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13158 | | Descriptor: | 1,2-ETHANEDIOL, 7-[2-[2,4-bis(fluoranyl)phenoxy]-5-(2-oxidanylpropan-2-yl)phenyl]-2-[4-(2-hydroxyethyloxy)-3,5-dimethyl-phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WLN
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13153 | | Descriptor: | 2-(2-cyclopentyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
