6KPG
 
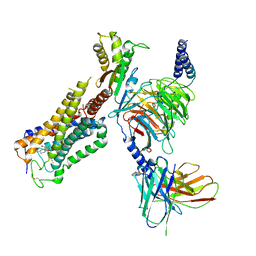 | | Cryo-EM structure of CB1-G protein complex | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hua, T, Li, X.T, Wu, L.J, Makriyannis, A, Wang, Y.X, Shen, L, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
5TGZ
 
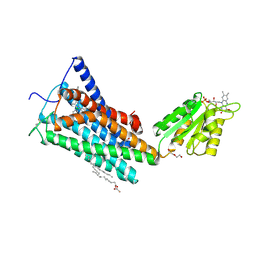 | | Crystal Structure of the Human Cannabinoid Receptor CB1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-[4-[2-(2,4-dichlorophenyl)-4-methyl-5-(piperidin-1-ylcarbamoyl)pyrazol-3-yl]phenyl]but-3-ynyl nitrate, Cannabinoid receptor 1,Flavodoxin,Cannabinoid receptor 1, ... | | Authors: | Hua, T, Vemuri, K, Pu, M, Qu, L, Han, G.W, Wu, Y, Zhao, S, Shui, W, Li, S, Korde, A, Laprairie, R.B, Stahl, E.L, Ho, J.H, Zvonok, N, Zhou, H, Kufareva, I, Wu, B, Zhao, Q, Hanson, M.A, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2016-09-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Human Cannabinoid Receptor CB1.
Cell, 167, 2016
|
|
5XR8
 
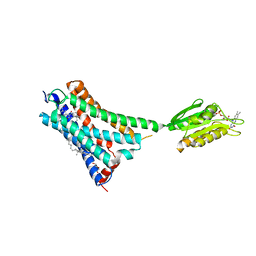 | | Crystal structure of the human CB1 in complex with agonist AM841 | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, CHOLESTEROL, Cannabinoid receptor 1,Flavodoxin,Cannabinoid receptor 1, ... | | Authors: | Hua, T, Vemuri, K, Nikas, P.S, Laprairie, R.B, Wu, Y, Qu, L, Pu, M, Korde, A, Shan, J, Ho, J.H, Han, G.W, Ding, K, Li, X, Liu, H, Hanson, M.A, Zhao, S, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structures of agonist-bound human cannabinoid receptor CB1.
Nature, 547, 2017
|
|
5XRA
 
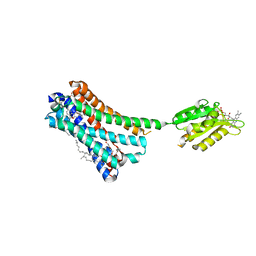 | | Crystal structure of the human CB1 in complex with agonist AM11542 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6aR,10aR)-3-(8-bromanyl-2-methyl-octan-2-yl)-6,6,9-trimethyl-6a,7,10,10a-tetrahydrobenzo[c]chromen-1-ol, CHOLESTEROL, ... | | Authors: | Hua, T, Vemuri, K, Nikas, P.S, Laprairie, R.B, Wu, Y, Qu, L, Pu, M, Korde, A, Shan, J, Ho, J.H, Han, G.W, Ding, K, Li, X, Liu, H, Hanson, M.A, Zhao, S, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2017-06-08 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of agonist-bound human cannabinoid receptor CB1
Nature, 547, 2017
|
|
4FC7
 
 | | Studies on DCR shed new light on peroxisomal beta-oxidation: Crystal structure of the ternary complex of pDCR | | Descriptor: | COENZYME A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Peroxisomal 2,4-dienoyl-CoA reductase | | Authors: | Hua, T, Wu, D, Wang, J, Shaw, N, Liu, Z.-J. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Studies of human 2,4-dienoyl CoA reductase shed new light on peroxisomal beta-oxidation of unsaturated fatty acids
J.Biol.Chem., 287, 2012
|
|
4FC6
 
 | | Studies on DCR shed new light on peroxisomal beta-oxidation: Crystal structure of the ternary complex of pDCR | | Descriptor: | HEXANOYL-COENZYME A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Peroxisomal 2,4-dienoyl-CoA reductase | | Authors: | Hua, T, Wu, D, Wang, J, Shaw, N, Liu, Z.-J. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Studies of human 2,4-dienoyl CoA reductase shed new light on peroxisomal beta-oxidation of unsaturated fatty acids
J.Biol.Chem., 287, 2012
|
|
6L82
 
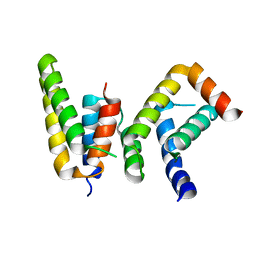 | | Crystal structure of Chaetomium GCP5 N-terminus and Mozart1 | | Descriptor: | Mozart1, Spindle pole body component | | Authors: | Huang, T.L, Wang, H.J, Wang, S.W, Hsia, K.C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.24103618 Å) | | Cite: | Promiscuous Binding of Microprotein Mozart1 to gamma-Tubulin Complex Mediates Specific Subcellular Targeting to Control Microtubule Array Formation.
Cell Rep, 31, 2020
|
|
6M33
 
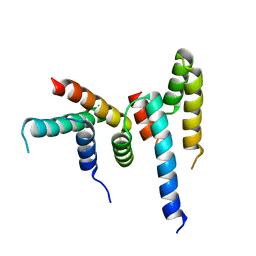 | |
4DNS
 
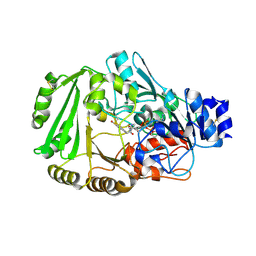 | | Crystal structure of Bermuda grass isoallergen BG60 provides insight into the various cross-allergenicity of the pollen group 4 allergens | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FAD-linked oxidoreductase BG60, ... | | Authors: | Huang, T.H, Peng, H.J, Su, S.N, Liaw, S.H. | | Deposit date: | 2012-02-08 | | Release date: | 2012-12-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Various cross-reactivity of the grass pollen group 4 allergens: crystallographic study of the Bermuda grass isoallergen Cyn d 4.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2MI8
 
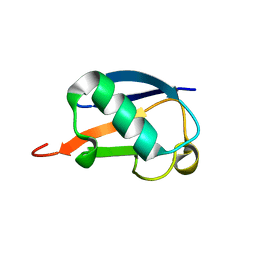 | |
6L81
 
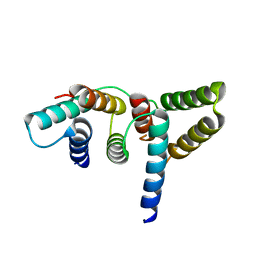 | | Crystal structure of Homo sapiens GCP5 N-terminus and Mozart1 | | Descriptor: | Gamma-tubulin complex component 5, Mitotic-spindle organizing protein 1 | | Authors: | Huang, T.L, Wang, H.J, Wang, S.W, Hsia, K.C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.19651 Å) | | Cite: | Promiscuous Binding of Microprotein Mozart1 to gamma-Tubulin Complex Mediates Specific Subcellular Targeting to Control Microtubule Array Formation.
Cell Rep, 31, 2020
|
|
8TPE
 
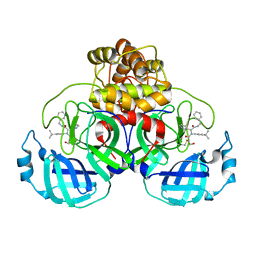 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | N-[(1R)-2-(benzylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)-3-hydroxypropanamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8TPG
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | (3R)-N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-3-hydroxybutanamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8TPB
 
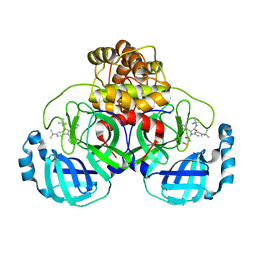 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)-2-chloroacetamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8TPH
 
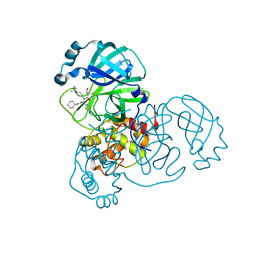 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | (3R)-N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-3-hydroxybutanamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8TPI
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-2-hydroxy-2-methylpropanamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8TPD
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | 3C-like proteinase nsp5, N-[(1R)-2-(benzylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-[3-(2-chloroacetamido)phenyl]furan-2-carboxamide | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8TPF
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-3-hydroxypropanamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8TPC
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | N-[(1R)-2-(benzylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-[4-(2-chloroacetamido)phenyl]furan-2-carboxamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
6L80
 
 | | Crystal structure of pombe Mod21 N-terminus and Mozart1 | | Descriptor: | Gamma-tubulin complex subunit mod21, Mitotic-spindle organizing protein 1 | | Authors: | Huang, T.L, Wang, H.J, Wang, S.W, Hsia, K.C. | | Deposit date: | 2019-11-03 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.00049758 Å) | | Cite: | Promiscuous Binding of Microprotein Mozart1 to gamma-Tubulin Complex Mediates Specific Subcellular Targeting to Control Microtubule Array Formation.
Cell Rep, 31, 2020
|
|
6L7R
 
 | | Crystal structure of Chaetomium GCP3 N-terminus and Mozart1 | | Descriptor: | Mozart1, Putative spindle pole body component alp6 protein | | Authors: | Huang, T.L, Wang, H.J, Hsia, K.C. | | Deposit date: | 2019-11-02 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8481313 Å) | | Cite: | Promiscuous Binding of Microprotein Mozart1 to gamma-Tubulin Complex Mediates Specific Subcellular Targeting to Control Microtubule Array Formation.
Cell Rep, 31, 2020
|
|
3HJ6
 
 | | Structure of Halothermothrix orenii fructokinase (FRK) | | Descriptor: | Fructokinase | | Authors: | Chua, T.K, Seetharaman, J, Kasprzak, J.M, Ng, C, Patel, B.K, Love, C, Bujnicki, J.M, Sivaraman, J. | | Deposit date: | 2009-05-21 | | Release date: | 2010-06-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a fructokinase homolog from Halothermothrix orenii
J.Struct.Biol., 171, 2010
|
|
8XQL
 
 | | Structure of human class T GPCR TAS2R14-miniGs/gust complex with Aristolochic acid A. | | Descriptor: | 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 2024
|
|
8XQR
 
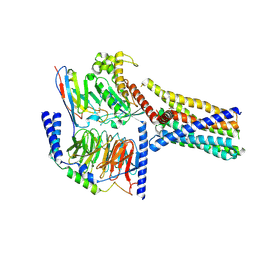 | | Structure 2 of human class T GPCR TAS2R14-miniGs/gust complex with Flufenamic acid. | | Descriptor: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 2024
|
|
8XQP
 
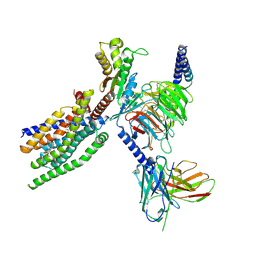 | | Structure of human class T GPCR TAS2R14-Gustducin complex with Aristolochic acid A. | | Descriptor: | 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 2024
|
|
