6L5L
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at apo state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5O
 
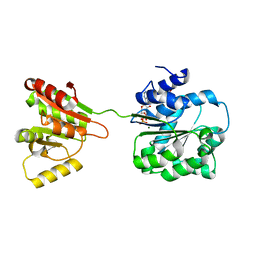 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-hydrolysis state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6S8Y
 
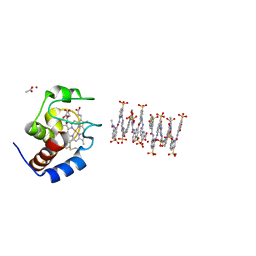 | | Crystal structure of cytochrome c in complex with a sulfonated quinoline-derived foldamer | | Descriptor: | 8-acetamido-2-[[2-[[2-[[2-[[2-[[2-[[2-[(2-carboxy-4-sulfonato-quinolin-8-yl)carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]quinoline-4-sulfonate, ACETATE ION, Cytochrome c iso-1, ... | | Authors: | Alex, J.M, Corvaglia, V, Hu, X, Engilberge, S, Huc, I, Crowley, P.B. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a protein-aromatic foldamer composite: macromolecular chiral resolution.
Chem.Commun.(Camb.), 55, 2019
|
|
5CB2
 
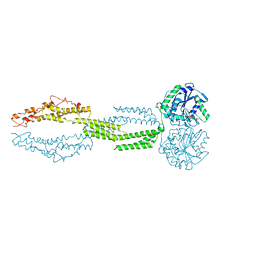 | | the structure of candida albicans Sey1p in complex with GMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Protein SEY1 | | Authors: | Yan, L, Sun, S, Wang, W, Shi, J, Hu, X, Wang, S, Rao, Z, Hu, J, Lou, Z. | | Deposit date: | 2015-06-30 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the yeast dynamin-like GTPase Sey1p provide insight into homotypic ER fusion
J.Cell Biol., 210, 2015
|
|
4X2A
 
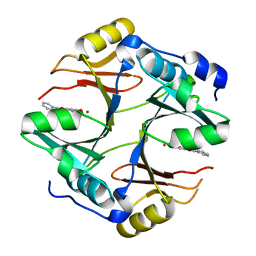 | | Crystal structure of mouse glyoxalase I complexed with baicalein | | Descriptor: | 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhang, H, Zhai, J, Zhang, L, Li, C, Zhao, Y, Hu, X. | | Deposit date: | 2014-11-26 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In Vitro Inhibition of Glyoxalase І by Flavonoids: New Insights from Crystallographic Analysis.
Curr Top Med Chem, 16, 2016
|
|
5O4N
 
 | | Apo HcgC from Methanococcus maripaludis soaked with SAH and pyridinol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-carboxy methyl-4-hydroxy-2-pyridinol, DIMETHYL SULFOXIDE, ... | | Authors: | Wagner, T, Bai, L, Xu, T, Hu, X, Ermler, U, Shima, S. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Water-Bridged H-Bonding Network Contributes to the Catalysis of the SAM-Dependent C-Methyltransferase HcgC.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5O4J
 
 | | HcgC from Methanococcus maripaludis cocrystallized with SAH and pyridinol | | Descriptor: | (3~{E})-3-[(~{E})-3-oxidanylprop-2-enoyl]iminopropanoic acid, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 6-carboxy methyl-4-hydroxy-2-pyridinol, ... | | Authors: | Wagner, T, Bai, L, Xu, T, Hu, X, Ermler, U, Shima, S. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Water-Bridged H-Bonding Network Contributes to the Catalysis of the SAM-Dependent C-Methyltransferase HcgC.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5O4M
 
 | | Fresh crystals of HcgC from Methanococcus maripaludis cocrystallized with SAH and pyridinol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-carboxy methyl-4-hydroxy-2-pyridinol, DIMETHYL SULFOXIDE, ... | | Authors: | Wagner, T, Bai, L, Xu, T, Hu, X, Ermler, U, Shima, S. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Water-Bridged H-Bonding Network Contributes to the Catalysis of the SAM-Dependent C-Methyltransferase HcgC.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5O4H
 
 | | HcgC from Methanococcus maripaludis cocrystallized with SAM and pyridinol | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ACETATE ION, HcgC, ... | | Authors: | Wagner, T, Bai, L, Xu, T, Hu, X, Ermler, U, Shima, S. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Water-Bridged H-Bonding Network Contributes to the Catalysis of the SAM-Dependent C-Methyltransferase HcgC.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
2KAX
 
 | | Solution structure and dynamics of S100A5 in the apo and Ca2+ -bound states | | Descriptor: | Protein S100-A5 | | Authors: | Bertini, I, Das Gupta, S, Hu, X, Karavelas, T, Luchinat, C, Parigi, G, Yuan, J, Structural Proteomics in Europe (SPINE), Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2008-11-17 | | Release date: | 2009-06-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of S100A5 in the apo and Ca2+-bound states
J.Biol.Inorg.Chem., 14, 2009
|
|
6U7V
 
 | | xRRM structure of spPof8 | | Descriptor: | NITRATE ION, Protein pof8 | | Authors: | Kim, J.-K, Hu, X, Yu, C, Jun, H.-I, Liu, J, Sankaran, B, Huang, L, Qiao, F. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Quality-Control Mechanism for Telomerase RNA Folding in the Cell.
Cell Rep, 33, 2020
|
|
5GNS
 
 | | Structures of human Mitofusin 1 provide insight into mitochondrial tethering | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Mitofusin-1 | | Authors: | Qi, Y, Yan, L, Yu, C, Guo, X, Zhou, X, Hu, X, Huang, X, Rao, Z, Lou, Z, Hu, J. | | Deposit date: | 2016-07-24 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structures of human Mitofusin 1 provide insight into mitochondrial tethering
To Be Published
|
|
4RVP
 
 | | Crystal structure of superoxide dismutase from sedum alfredii | | Descriptor: | ZINC ION, superoxide dismutase | | Authors: | Qiu, R, Li, C, Zhai, J, Tang, L, Zhang, H, Yuan, M, Hu, X. | | Deposit date: | 2014-11-27 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The positive effects of Cd and Cd-Zn relationship in the
Zn-related physiological processes involved in growth in the Zn/Cd hyperaccumulator Sedum alfredii
To be Published
|
|
6V6A
 
 | | Inhibitory scaffolding of the ancient MAPK, ERK7 | | Descriptor: | 1,2-ETHANEDIOL, Apical Cap Protein 9 (AC9), Mitogen-activated protein kinase | | Authors: | Dewangan, P.S, O'Shaughnessy, W.J, Back, P.S, Hu, X, Bradley, P.J, Reese, M.L. | | Deposit date: | 2019-12-04 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ancient MAPK ERK7 is regulated by an unusual inhibitory scaffold required forToxoplasmaapical complex biogenesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4XO7
 
 | | Crystal structure of human 3-alpha hydroxysteroid dehydrogenase type 3 in complex with NADP+, 5alpha-androstan-3,17-dione and (3beta, 5alpha)-3-hydroxyandrostan-17-one | | Descriptor: | 4-ANDROSTENE-3-17-DIONE, Aldo-keto reductase family 1 member C2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, B, Hu, X.-J, Lin, S.-X. | | Deposit date: | 2015-01-16 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Human 3 alpha-hydroxysteroid dehydrogenase type 3: structural clues of 5 alpha-DHT reverse binding and enzyme down-regulation decreasing MCF7 cell growth.
Biochem.J., 473, 2016
|
|
2XWT
 
 | | CRYSTAL STRUCTURE OF THE TSH RECEPTOR IN COMPLEX WITH A BLOCKING TYPE TSHR AUTOANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, THYROID BLOCKING HUMAN AUTOANTIBODY K1-70 HEAVY CHAIN, ... | | Authors: | Sanders, J, Sanders, P, Young, S, Kabelis, K, Baker, S, Sullivan, A, Evans, M, Clark, J, Wilmot, J, Hu, X, Roberts, E, Powell, M, Nunez Miguel, R, Furmaniak, J, Rees Smith, B. | | Deposit date: | 2010-11-05 | | Release date: | 2011-03-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Tsh Receptor (Tshr) Bound to a Blocking-Type Tshr Autoantibody.
J.Mol.Endocrinol., 46, 2011
|
|
4XO6
 
 | | Crystal structure of human 3-alpha hydroxysteroid dehydrogenase type 3 in complex with NADP+, 5alpha-androstan-3,17-dione and (3beta, 5alpha)-3-hydroxyandrostan-17-one | | Descriptor: | (3Beta,5alpha)-3-Hydroxyandrostan-17-one, 1,2-ETHANEDIOL, 5ALPHA-ANDROSTAN-3,17-DIONE, ... | | Authors: | Zhang, B, Hu, X.-J, Lin, S.-X. | | Deposit date: | 2015-01-16 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Human 3 alpha-hydroxysteroid dehydrogenase type 3: structural clues of 5 alpha-DHT reverse binding and enzyme down-regulation decreasing MCF7 cell growth.
Biochem.J., 473, 2016
|
|
2KAY
 
 | | Solution structure and dynamics of S100A5 in the Ca2+ -bound states | | Descriptor: | CALCIUM ION, Protein S100-A5 | | Authors: | Bertini, I, Das Gupta, S, Hu, X, Karavelas, T, Luchinat, C, Parigi, G, Yuan, J, Structural Proteomics in Europe (SPINE), Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2008-11-17 | | Release date: | 2009-06-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of S100A5 in the apo and Ca2+-bound states
J.Biol.Inorg.Chem., 14, 2009
|
|
1LGH
 
 | | CRYSTAL STRUCTURE OF THE LIGHT-HARVESTING COMPLEX II (B800-850) FROM RHODOSPIRILLUM MOLISCHIANUM | | Descriptor: | BACTERIOCHLOROPHYLL A, HEPTANE-1,2,3-TRIOL, LIGHT HARVESTING COMPLEX II, ... | | Authors: | Koepke, J, Hu, X, Schulten, K, Michel, H. | | Deposit date: | 1996-03-20 | | Release date: | 1997-06-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the light-harvesting complex II (B800-850) from Rhodospirillum molischianum.
Structure, 4, 1996
|
|
4U7N
 
 | | Inactive structure of histidine kinase | | Descriptor: | Histidine protein kinase sensor protein | | Authors: | Cai, Y, Hu, X, Sang, J. | | Deposit date: | 2014-07-31 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational dynamics of the essential sensor histidine kinase WalK.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
4U7O
 
 | | Active histidine kinase bound with ATP | | Descriptor: | AMP PHOSPHORAMIDATE, Histidine protein kinase sensor protein | | Authors: | Cai, Y, Hu, X, Sang, J. | | Deposit date: | 2014-07-31 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Conformational dynamics of the essential sensor histidine kinase WalK.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1X9Z
 
 | | Crystal structure of the MutL C-terminal domain | | Descriptor: | CHLORIDE ION, DNA mismatch repair protein mutL, GLYCEROL, ... | | Authors: | Guarne, A, Ramon-Maiques, S, Wolff, E.M, Ghirlando, R, Hu, X, Miller, J.H, Yang, W. | | Deposit date: | 2004-08-24 | | Release date: | 2004-10-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the MutL C-terminal domain: a model of intact MutL and its roles in mismatch repair
Embo J., 23, 2004
|
|
5HNU
 
 | | Crystal Structure of AKR1C3 complexed with octyl gallate | | Descriptor: | Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, octyl 3,4,5-trihydroxybenzoate | | Authors: | Li, C, Zhao, Y, Zhang, H, Hu, X. | | Deposit date: | 2016-01-18 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of AKR1C3 complexed with octyl gallte
To Be Published
|
|
4PV5
 
 | | Crystal structure of mouse glyoxalase I in complexed with 18-beta-glycyrrhetinic acid | | Descriptor: | (3BETA,5BETA,14BETA)-3-HYDROXY-11-OXOOLEAN-12-EN-29-OIC ACID, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhang, H, Zhai, J, Zhang, L.P, Zhao, Y.N, Li, C, Hu, X.P. | | Deposit date: | 2014-03-15 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for 18-beta-glycyrrhetinic acid as a novel non-GSH analog glyoxalase I inhibitor
Acta Pharmacol.Sin., 36, 2015
|
|
1YCL
 
 | | Crystal Structure of B. subtilis LuxS in Complex with a Catalytic 2-Ketone Intermediate | | Descriptor: | (S)-2-AMINO-4-[(2S,3R)-2,3,5-TRIHYDROXY-4-OXO-PENTYL]MERCAPTO-BUTYRIC ACID, COBALT (II) ION, S-ribosylhomocysteinase, ... | | Authors: | Rajan, R, Zhu, J, Hu, X, Pei, D, Bell, C.E. | | Deposit date: | 2004-12-22 | | Release date: | 2005-03-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of S-Ribosylhomocysteinase (LuxS) in Complex with a Catalytic 2-Ketone Intermediate.
Biochemistry, 44, 2005
|
|
