3LZ2
 
 | | STRUCTURE DETERMINATION OF TURKEY EGG WHITE LYSOZYME USING LAUE DIFFRACTION | | Descriptor: | TURKEY EGG WHITE LYSOZYME | | Authors: | Howell, P.L, Almo, S.C, Parsons, M.R, Hajdu, J, Petsko, G.A. | | Deposit date: | 1991-09-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure determination of turkey egg-white lysozyme using Laue diffraction data.
Acta Crystallogr.,Sect.B, 48, 1992
|
|
1TEW
 
 | |
5UVR
 
 | |
4NK6
 
 | |
4OZX
 
 | |
4OZZ
 
 | |
4OZV
 
 | | Crystal Structure of the periplasmic alginate lyase AlgL | | Descriptor: | Alginate lyase, beta-D-mannopyranuronic acid | | Authors: | Howell, P.L, Wolfram, F, Robinson, H, Arora, K. | | Deposit date: | 2014-02-19 | | Release date: | 2015-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | The Pseudomonas aeruginosa homeostasis enzyme AlgL clears the periplasmic space of accumulated alginate during polymer biosynthesis.
J.Biol.Chem., 298, 2022
|
|
4OZW
 
 | |
4NK8
 
 | |
4OZY
 
 | |
5WFT
 
 | | PelB 319-436 from Pseudomonas aeruginosa PAO1 | | Descriptor: | PelB | | Authors: | Marmont, L.S, Howell, P.L. | | Deposit date: | 2017-07-12 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.821 Å) | | Cite: | PelA and PelB proteins form a modification and secretion complex essential for Pel polysaccharide-dependent biofilm formation in Pseudomonas aeruginosa.
J. Biol. Chem., 292, 2017
|
|
9CGY
 
 | |
4DN0
 
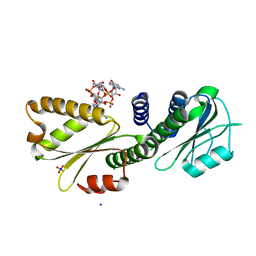 | | PelD 156-455 from Pseudomonas aeruginosa PA14 in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Putative uncharacterized protein pelD, SODIUM ION, ... | | Authors: | Whitney, J.C, Colvin, K.M, Marmont, L.S, Robinson, H, Parsek, M.R, Howell, P.L. | | Deposit date: | 2012-02-08 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Cytoplasmic Region of PelD, a Degenerate Diguanylate Cyclase Receptor That Regulates Exopolysaccharide Production in Pseudomonas aeruginosa.
J.Biol.Chem., 287, 2012
|
|
7ULA
 
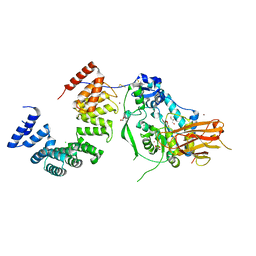 | | Structure of the Pseudomonas putida AlgKX modification and secretion complex | | Descriptor: | Alginate biosynthesis protein AlgK, Alginate biosynthesis protein AlgX, CHLORIDE ION, ... | | Authors: | Gheorghita, A.A, Li, E.Y, Pfoh, R, Howell, P.L. | | Deposit date: | 2022-04-04 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure of the AlgKX modification and secretion complex required for alginate production and biofilm attachment in Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
2PTQ
 
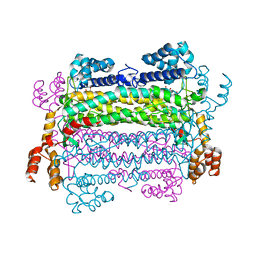 | |
3FZ2
 
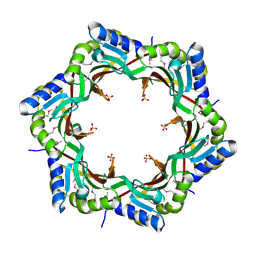 | | Crystal structure of the tail terminator protein from phage lambda (gpU-D74A) | | Descriptor: | Minor tail protein U, SULFATE ION | | Authors: | Pell, L.G, Liu, A, Edmonds, E, Donaldson, L.W, Howell, P.L, Davidson, A.R. | | Deposit date: | 2009-01-23 | | Release date: | 2009-05-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages.
J.Mol.Biol., 389, 2009
|
|
3FZB
 
 | | Crystal structure of the tail terminator protein from phage lambda (gpU-WT) | | Descriptor: | Minor tail protein U, SULFATE ION | | Authors: | Pell, L.G, Liu, A, Edmonds, E, Donaldson, L.W, Howell, P.L, Davidson, A.R. | | Deposit date: | 2009-01-24 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages.
J.Mol.Biol., 389, 2009
|
|
4MBQ
 
 | | TPR3 of FimV from P. aeruginosa (PAO1) | | Descriptor: | Motility protein FimV | | Authors: | Nguyen, Y, Zhang, K, Daniel-Ivad, M, Robinson, H, Wolfram, F, Sugiman-Marangos, S.N, Junop, M.S, Burrows, L.L, Howell, P.L. | | Deposit date: | 2013-08-19 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of TPR2 from FimV
To be Published
|
|
4MAL
 
 | | TPR3 of FimV from P. aeruginosa (PAO1) | | Descriptor: | Motility protein FimV | | Authors: | Nguyen, Y, Zhang, K, Daniel-Ivad, M, Sugiman-Marangos, S.N, Junop, M.S, Burrows, L.L, Howell, P.L. | | Deposit date: | 2013-08-16 | | Release date: | 2014-08-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of TPR2 from FimV
To be Published
|
|
3LGS
 
 | |
1Z5P
 
 | | Crystal structure of MTA/AdoHcy nucleosidase with a ligand-free purine binding site | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Lee, J.E, Smith, G.D, Horvatin, C, Huang, D.J.T, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2005-03-18 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural snapshots of MTA/AdoHcy nucleosidase along the reaction coordinate provide insights into enzyme and nucleoside flexibility during catalysis
J.Mol.Biol., 352, 2005
|
|
1Z5O
 
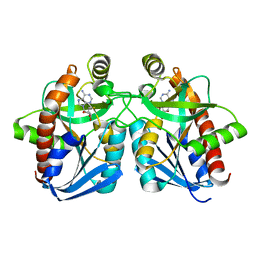 | | Crystal structure of MTA/AdoHcy nucleosidase Asp197Asn mutant complexed with 5'-methylthioadenosine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Smith, G.D, Horvatin, C, Huang, D.J.T, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2005-03-18 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural snapshots of MTA/AdoHcy nucleosidase along the reaction coordinate provide insights into enzyme and nucleoside flexibility during catalysis
J.Mol.Biol., 352, 2005
|
|
1Z5N
 
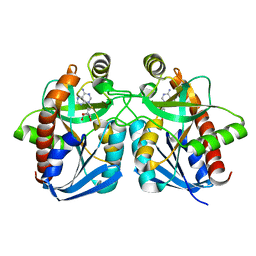 | | Crystal structure of MTA/AdoHcy nucleosidase Glu12Gln mutant complexed with 5-methylthioribose and adenine | | Descriptor: | 5-S-methyl-5-thio-alpha-D-ribofuranose, ADENINE, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Smith, G.D, Horvatin, C, Huang, D.J.T, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2005-03-18 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural snapshots of MTA/AdoHcy nucleosidase along the reaction coordinate provide insights into enzyme and nucleoside flexibility during catalysis
J.Mol.Biol., 352, 2005
|
|
1Y6Q
 
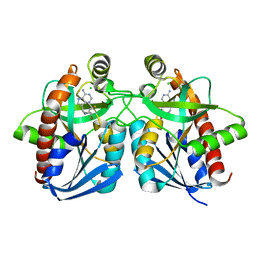 | | Cyrstal structure of MTA/AdoHcy nucleosidase complexed with MT-DADMe-ImmA | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, CHLORIDE ION, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Singh, V, Evans, G.B, Tyler, P.C, Furneaux, R.H, Cornell, K.A, Riscoe, M.K, Schramm, V.L, Howell, P.L. | | Deposit date: | 2004-12-06 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural rationale for the affinity of pico- and femtomolar transition state analogues of Escherichia coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
J.Biol.Chem., 280, 2005
|
|
1Y6R
 
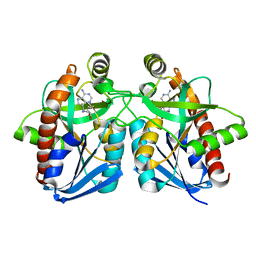 | | Crystal structure of MTA/AdoHcy nucleosidase complexed with MT-ImmA. | | Descriptor: | (3S,4R)-2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-[(METHYLSULFANYL)METHYL]PYRROLIDINE-3,4-DIOL, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Singh, V, Evans, G.B, Tyler, P.C, Furneaux, R.H, Cornell, K.A, Riscoe, M.K, Schramm, V.L, Howell, P.L. | | Deposit date: | 2004-12-06 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural rationale for the affinity of pico- and femtomolar transition state analogues of Escherichia coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
J.Biol.Chem., 280, 2005
|
|
