6K2U
 
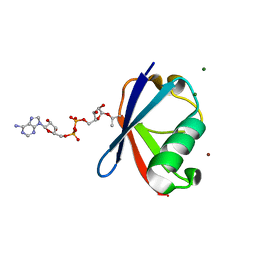 | | Crystal structure of Thr66 ADP-ribosylated ubiquitin | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, MAGNESIUM ION, Polyubiquitin-C, ... | | Authors: | Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2019-05-15 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Threonine ADP-Ribosylation of Ubiquitin by a Bacterial Effector Family Blocks Host Ubiquitination.
Mol.Cell, 78, 2020
|
|
5ZNS
 
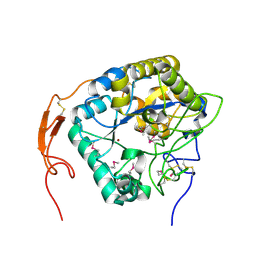 | | Insect chitin deacetylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ZINC ION, ... | | Authors: | Liu, L, Zhou, Y, Yang, Q. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Structural and biochemical insights into the catalytic mechanisms of two insect chitin deacetylases of the carbohydrate esterase 4 family.
J. Biol. Chem., 294, 2019
|
|
4M7A
 
 | | Crystal structure of Lsm2-8 complex bound to the 3' end sequence of U6 snRNA | | Descriptor: | U6 snRNA, U6 snRNA-associated Sm-like protein LSm2, U6 snRNA-associated Sm-like protein LSm3, ... | | Authors: | Zhou, L, Hang, J, Zhou, Y, Wan, R, Lu, G, Yan, C, Shi, Y. | | Deposit date: | 2013-08-12 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.781 Å) | | Cite: | Crystal structures of the Lsm complex bound to the 3' end sequence of U6 small nuclear RNA.
Nature, 506, 2014
|
|
6AF0
 
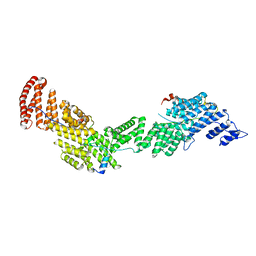 | | Structure of Ctr9, Paf1 and Cdc73 ternary complex from Myceliophthora thermophila | | Descriptor: | Cdc73 protein, Ctr9 protein, Paf1 protein | | Authors: | Wang, Z, Deng, P, Zhou, Y. | | Deposit date: | 2018-08-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Transcriptional elongation factor Paf1 core complex adopts a spirally wrapped solenoidal topology.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5ZNT
 
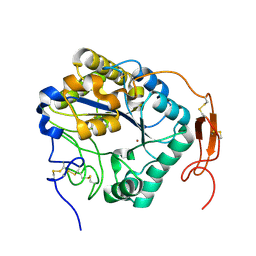 | | Insect chitin deacetylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ZINC ION, ... | | Authors: | Liu, L, Zhou, Y, Yang, Q. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structural and biochemical insights into the catalytic mechanisms of two insect chitin deacetylases of the carbohydrate esterase 4 family.
J. Biol. Chem., 294, 2019
|
|
6B7N
 
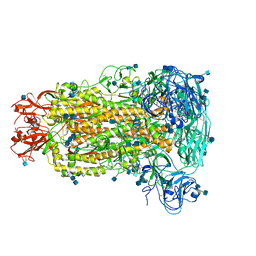 | | Cryo-electron microscopy structure of porcine delta coronavirus spike protein in the pre-fusion state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shang, J, Zheng, Y, Yang, Y, Liu, C, Geng, Q, Tai, W, Du, L, Zhou, Y, Zhang, W, Li, F. | | Deposit date: | 2017-10-04 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-Electron Microscopy Structure of Porcine Deltacoronavirus Spike Protein in the Prefusion State
J. Virol., 92, 2018
|
|
3O0J
 
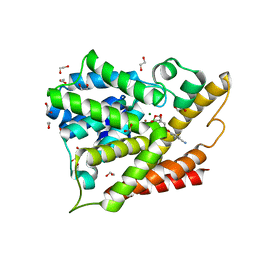 | | PDE4B In complex with ligand an2898 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(1-hydroxy-1,3-dihydro-2,1-benzoxaborol-5-yl)oxy]benzene-1,2-dicarbonitrile, MAGNESIUM ION, ... | | Authors: | Alley, M.R.K, Zhou, Y. | | Deposit date: | 2010-07-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Boron-based phosphodiesterase inhibitors show novel binding of boron to PDE4 bimetal center.
Febs Lett., 586, 2012
|
|
6ACV
 
 | | the solution NMR structure of MBD domain | | Descriptor: | Methyl-CpG-binding domain-containing protein 11 | | Authors: | Li, S.L, Feng, Y.Y, Zhou, Y, Ding, Y.M, Liu, K, Nie, Y, Li, F, Yang, Y.Y. | | Deposit date: | 2018-07-27 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | the solution NMR structure of MBD domains
To Be Published
|
|
3WNV
 
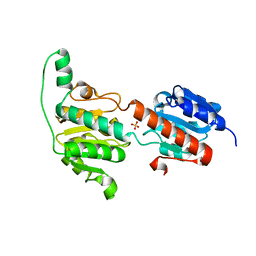 | | Crystal structure of a glyoxylate reductase from Paecilomyes thermophila | | Descriptor: | SULFATE ION, glyoxylate reductase | | Authors: | Duan, X, Hu, S, Zhou, P, Zhou, Y, Jiang, Z. | | Deposit date: | 2013-12-17 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization and crystal structure of a first fungal glyoxylate reductase from Paecilomyes thermophila
Enzyme.Microb.Technol., 60, 2014
|
|
5Y1B
 
 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with a berberine derivative (SYSU-00679) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-3'-quinolinium propylberberine, Beta-hexosaminidase | | Authors: | Duan, Y.W, Liu, T, Zhou, Y, Tang, J.Y, Li, M, Yang, Q. | | Deposit date: | 2017-07-20 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.207 Å) | | Cite: | Glycoside hydrolase family 18 and 20 enzymes are novel targets of the traditional medicine berberine.
J. Biol. Chem., 293, 2018
|
|
7F5G
 
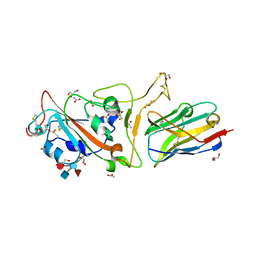 | | The crystal structure of RBD-Nanobody complex, DL4 (SA4) | | Descriptor: | ACETATE ION, GLYCEROL, Nanobody DL4, ... | | Authors: | Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Isolation, characterization, and structure-based engineering of a neutralizing nanobody against SARS-CoV-2.
Int.J.Biol.Macromol., 209, 2022
|
|
7F5H
 
 | | The crystal structure of RBD-Nanobody complex, DL28 (SC4) | | Descriptor: | GLYCEROL, Nanobody DL28, PHOSPHATE ION, ... | | Authors: | Luo, Z.P, Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Characterization of a Neutralizing Nanobody With Broad Activity Against SARS-CoV-2 Variants.
Front Microbiol, 13, 2022
|
|
6IGY
 
 | |
6JAW
 
 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 in complex with a napthalimide derivative | | Descriptor: | 2-[3-(morpholin-4-yl)propyl]-1H-benzo[de]isoquinoline-1,3(2H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, Group II chitinase | | Authors: | Chen, W, Zhou, Y, Yang, Q. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Structural dissection reveals a general mechanistic principle for group II chitinase (ChtII) inhibition.
J.Biol.Chem., 294, 2019
|
|
6JAV
 
 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 in complex with a piperidine-thienopyridine derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(4-chlorophenyl)methyl]sulfanyl}-7-methyl-N-(prop-2-en-1-yl)-7,8-dihydropyrido[4',3':4,5]thieno[2,3-d]pyrimidin-4-amine, Group II chitinase | | Authors: | Chen, W, Zhou, Y, Yang, Q. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.437 Å) | | Cite: | Structural dissection reveals a general mechanistic principle for group II chitinase (ChtII) inhibition.
J.Biol.Chem., 294, 2019
|
|
6JAY
 
 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 in complex with a dipyrido-pyrimidine derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-1-[(furan-2-yl)methyl]-5-oxo-3-({[(2S)-oxolan-2-yl]methyl}carbamoyl)-5H-dipyrido[1,2-a:2',3'-d]pyrimidin-1-ium, Group II chitinase | | Authors: | Chen, W, Zhou, Y, Yang, Q. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Structural dissection reveals a general mechanistic principle for group II chitinase (ChtII) inhibition.
J.Biol.Chem., 294, 2019
|
|
6JAX
 
 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 in complex with chitooctaose [(GlcN)8] | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, Group II chitinase | | Authors: | Chen, W, Zhou, Y, Yang, Q. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural dissection reveals a general mechanistic principle for group II chitinase (ChtII) inhibition.
J.Biol.Chem., 294, 2019
|
|
5Y2A
 
 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 2 | | Descriptor: | alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, insect group II chitinase | | Authors: | Chen, W, Qu, M.B, Zhou, Y, Yang, Q. | | Deposit date: | 2017-07-24 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of group II chitinase (ChtII) catalysis completes the puzzle of chitin hydrolysis in insects
J. Biol. Chem., 293, 2018
|
|
5Y29
 
 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, insect group II chitinase | | Authors: | Chen, W, Qu, M.B, Zhou, Y, Yang, Q. | | Deposit date: | 2017-07-24 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of group II chitinase (ChtII) catalysis completes the puzzle of chitin hydrolysis in insects
J. Biol. Chem., 293, 2018
|
|
5Z34
 
 | |
3RXR
 
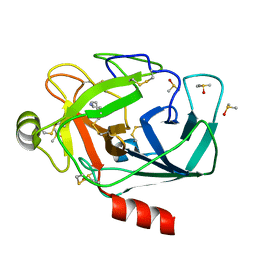 | | Crystal structure of Trypsin complexed with cycloheptanamine (F01 and F03, cocktail experiment) | | Descriptor: | CALCIUM ION, Cationic trypsin, DIMETHYL SULFOXIDE, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXS
 
 | | Crystal structure of Trypsin complexed with (3-methoxyphenyl)methanamine (F04 and A06, cocktail experiment) | | Descriptor: | 1-(3-methoxyphenyl)methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
5ZUT
 
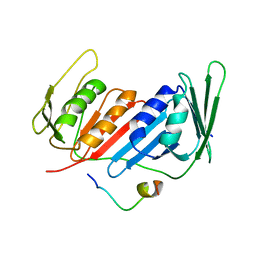 | | Crystal Structure of Yeast PCNA in Complex with N24 Peptide | | Descriptor: | N24, Proliferating cell nuclear antigen | | Authors: | Cheng, X.Y, Kuang, X.L, Zhou, Y, Xia, X.M, SU, Z.D. | | Deposit date: | 2018-05-08 | | Release date: | 2018-05-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal Structure of Yeast PCNA in Complex with N24 Peptide
To Be Published
|
|
5Y2B
 
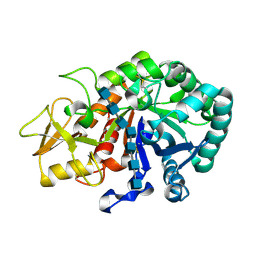 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 in complex with HEPTA-N-ACETYLCHITOOCTAOSE (NAG)7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, insect group II chitinase | | Authors: | Chen, W, Qu, M.B, Zhou, Y, Yang, Q. | | Deposit date: | 2017-07-24 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of group II chitinase (ChtII) catalysis completes the puzzle of chitin hydrolysis in insects
J. Biol. Chem., 293, 2018
|
|
5Y2C
 
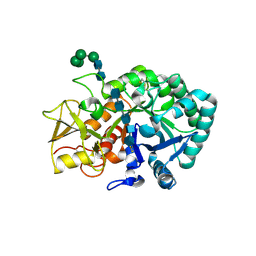 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 2 E2180L mutant in complex with PENTA-N-ACETYLCHITOOCTAOSE (NAG)5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, insect group II chitinase | | Authors: | Chen, W, Qu, M.B, Zhou, Y, Yang, Q. | | Deposit date: | 2017-07-24 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural analysis of group II chitinase (ChtII) catalysis completes the puzzle of chitin hydrolysis in insects
J. Biol. Chem., 293, 2018
|
|
