3IS1
 
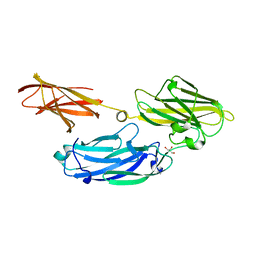 | | Crystal structure of functional region of UafA from Staphylococcus saprophyticus in C2 form at 2.45 angstrom resolution | | Descriptor: | GLYCEROL, Uro-adherence factor A | | Authors: | Tanaka, Y, Matsuoka, E, Shouji, Y, Kuroda, M, Tanaka, I, Yao, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the functional region of Uro-adherence factor A from Staphylococcus saprophyticus reveals participation of the B domain in ligand binding
Protein Sci., 20, 2011
|
|
3IS0
 
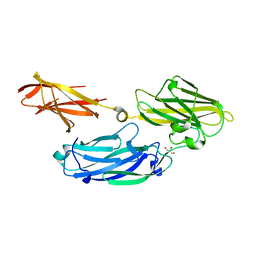 | | Crystal structure of functional region of UafA from Staphylococcus saprophyticus in the presence of cholesterol | | Descriptor: | GLYCEROL, Uro-adherence factor A | | Authors: | Tanaka, Y, Shouji, Y, Matsuoka, E, Kuroda, M, Tanaka, I, Yao, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Functional Region of UafA from Staphylococcus saprophyticus
To be Published
|
|
3IRP
 
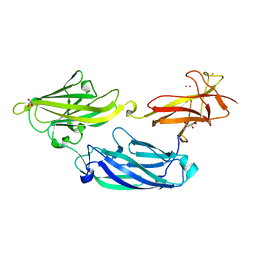 | | Crystal structure of functional region of UafA from Staphylococcus saprophyticus at 1.50 angstrom resolution | | Descriptor: | GLYCEROL, POTASSIUM ION, Uro-adherence factor A | | Authors: | Tanaka, Y, Shouji, Y, Matsuoka, E, Kuroda, M, Tanaka, I, Yao, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the functional region of Uro-adherence factor A from Staphylococcus saprophyticus reveals participation of the B domain in ligand binding
Protein Sci., 20, 2011
|
|
3IRZ
 
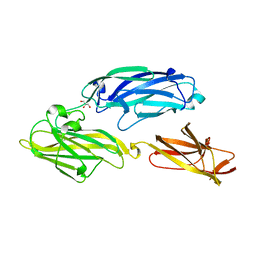 | | Crystal structure of functional region of UafA from Staphylococcus saprophyticus in P212121 form | | Descriptor: | GLYCEROL, Uro-adherence factor A | | Authors: | Tanaka, Y, Shouji, Y, Matsuoka, E, Kuroda, M, Tanaka, I, Yao, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the functional region of Uro-adherence factor A from Staphylococcus saprophyticus reveals participation of the B domain in ligand binding
Protein Sci., 20, 2011
|
|
2JXG
 
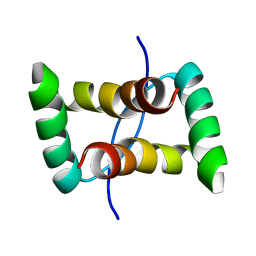 | |
2JXI
 
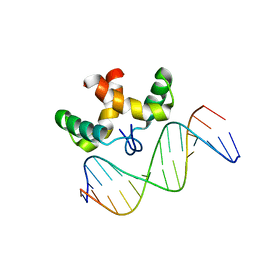 | | Solution structure of the DNA-binding domain of Pseudomonas putida Proline utilization A (putA) bound to GTTGCA DNA sequence | | Descriptor: | DNA (5'-D(*DAP*DAP*DAP*DGP*DGP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DC)-3'), DNA (5'-D(*DGP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DAP*DCP*DCP*DTP*DTP*DT)-3'), Proline dehydrogenase | | Authors: | Halouska, S, Zhou, Y, Becker, D, Powers, R. | | Deposit date: | 2007-11-19 | | Release date: | 2008-10-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Pseudomonas putida protein PpPutA45 and its DNA complex
Proteins, 75, 2008
|
|
7YFN
 
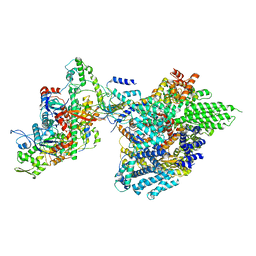 | | Core module of the NuA4 complex in S. cerevisiae | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ARP4 isoform 1, Actin, ... | | Authors: | Ji, L.T, Zhao, L.X, Xu, K, Gao, H.H, Zhou, Y, Kornberg, R.D, Zhang, H.Q. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the NuA4 histone acetyltransferase complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YFP
 
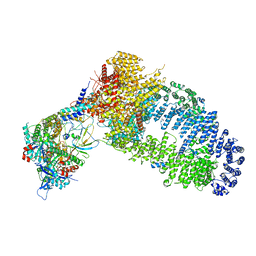 | | The NuA4 histone acetyltransferase complex from S. cerevisiae | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ARP4 isoform 1, Actin, ... | | Authors: | Ji, L.T, Zhao, L.X, Xu, K, Gao, H.H, Zhou, Y, Kornberg, R.D, Zhang, H.Q. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the NuA4 histone acetyltransferase complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3ZJT
 
 | | Ternary complex of E.coli leucyl-tRNA synthetase, tRNA(Leu)574 and the benzoxaborole AN3017 in the editing conformation | | Descriptor: | LEUCYL-TRNA SYNTHETASE, MAGNESIUM ION, TRNALEU5 UAA ISOACCEPTOR | | Authors: | Cusack, S, Palencia, A, Crepin, T, Hernandez, V, Akama, T, Baker, S.J, Bu, W, Feng, L, Freund, Y.R, Liu, L, Meewan, M, Mohan, M, Mao, W, Rock, F.L, Sexton, H, Sheoran, A, Zhang, Y, Zhang, Y, Zhou, Y, Nieman, J.A, Anugula, M.R, Keramane, E.M, Savariraj, K, Reddy, D.S, Sharma, R, Subedi, R, Singh, R, OLeary, A, Simon, N.L, DeMarsh, P.L, Mushtaq, S, Warner, M, Livermore, D.M, Alley, M.R.K, Plattner, J.J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Novel Class of Boron-Based Antibacterials with Activity Against Gram-Negative Bacteria.
Antimicrob.Agents Chemother., 57, 2013
|
|
3ZJU
 
 | | Ternary complex of E .coli leucyl-tRNA synthetase, tRNA(Leu) and the benzoxaborole AN3016 in the editing conformation | | Descriptor: | LEUCYL-TRNA SYNTHETASE, MAGNESIUM ION, TRNALEU5 UAA ISOACCEPTOR | | Authors: | Cusack, S, Palencia, A, Crepin, T, Hernandez, V, Akama, T, Baker, S.J, Bu, W, Feng, L, Freund, Y.R, Liu, L, Meewan, M, Mohan, M, Mao, W, Rock, F.L, Sexton, H, Sheoran, A, Zhang, Y, Zhang, Y, Zhou, Y, Nieman, J.A, Anugula, M.R, Keramane, E.M, Savariraj, K, Reddy, D.S, Sharma, R, Subedi, R, Singh, R, OLeary, A, Simon, N.L, DeMarsh, P.L, Mushtaq, S, Warner, M, Livermore, D.M, Alley, M.R.K, Plattner, J.J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Novel Class of Boron-Based Antibacterials with Activity Against Gram-Negative Bacteria.
Antimicrob.Agents Chemother., 57, 2013
|
|
3ZJV
 
 | | Ternary complex of E .coli leucyl-tRNA synthetase, tRNA(Leu) and the benzoxaborole AN3213 in the editing conformation | | Descriptor: | LEUCINE--TRNA LIGASE, TRNALEU5 UAA ISOACCEPTOR | | Authors: | Cusack, S, Palencia, A, Crepin, T, Hernandez, V, Akama, T, Baker, S.J, Bu, W, Feng, L, Freund, Y.R, Liu, L, Meewan, M, Mohan, M, Mao, W, Rock, F.L, Sexton, H, Sheoran, A, Zhang, Y, Zhang, Y, Zhou, Y, Nieman, J.A, Anugula, M.R, Keramane, E.M, Savariraj, K, Reddy, D.S, Sharma, R, Subedi, R, Singh, R, OLeary, A, Simon, N.L, DeMarsh, P.L, Mushtaq, S, Warner, M, Livermore, D.M, Alley, M.R.K, Plattner, J.J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery of a Novel Class of Boron-Based Antibacterials with Activity Against Gram-Negative Bacteria.
Antimicrob.Agents Chemother., 57, 2013
|
|
2AY0
 
 | | Structure of the Lys9Met mutant of the E. coli Proline Utilization A (PutA) DNA-binding domain. | | Descriptor: | Bifunctional putA protein, CHLORIDE ION | | Authors: | Larson, J.D, Schuermann, J.P, Zhou, Y, Jenkins, J.L, Becker, D.F, Tanner, J.J. | | Deposit date: | 2005-09-06 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the DNA-binding domain of Escherichia coli proline utilization A flavoprotein and analysis of the role of Lys9 in DNA recognition.
Protein Sci., 15, 2006
|
|
3WMC
 
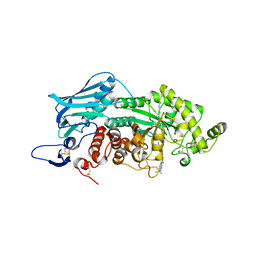 | | Crystal structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with naphthalimide derivative Q2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(dimethylamino)-2-(2-{[(5-methyl-1,3,4-thiadiazol-2-yl)methyl]amino}ethyl)-1H-benzo[de]isoquinoline-1,3(2H)-dione, Beta-hexosaminidase | | Authors: | Chen, L, Zhou, Y, Chen, L, Yang, Q. | | Deposit date: | 2013-11-16 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | A crystal structure-guided rational design switching non-carbohydrate inhibitors' specificity between two beta-GlcNAcase homologs
Sci Rep, 4, 2014
|
|
3WQW
 
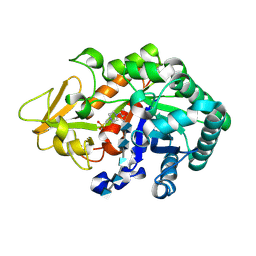 | | Crystal structure of Ostrinia furnacalis Group I chitinase catalytic domain in complex with a(GlcN)6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Chen, L, Zhou, Y, Yang, Q. | | Deposit date: | 2014-02-03 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fully deacetylated chitooligosaccharides act as efficient glycoside hydrolase family 18 chitinase inhibitors.
J.Biol.Chem., 289, 2014
|
|
2DWE
 
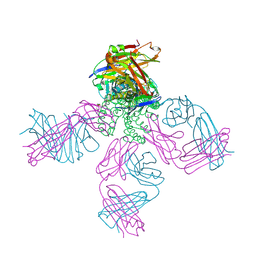 | | Crystal structure of KcsA-FAB-TBA complex in Rb+ | | Descriptor: | (2S)-3-HYDROXY-2-(NONANOYLOXY)PROPYL LAURATE, ANTIBODY FAB HEAVY CHAIN, ANTIBODY FAB LIGHT CHAIN, ... | | Authors: | Yohannan, S, Zhou, Y. | | Deposit date: | 2006-08-10 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic Study of the Tetrabutylammonium Block to the KcsA K(+) Channel
J.Mol.Biol., 366, 2007
|
|
3WQV
 
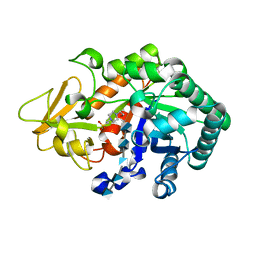 | | Crystal structure of Ostrinia furnacalis Group I chitinase catalytic domain in complex with a(GlcN)5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Chen, L, Zhou, Y, Yang, Q. | | Deposit date: | 2014-02-03 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Fully deacetylated chitooligosaccharides act as efficient glycoside hydrolase family 18 chitinase inhibitors.
J.Biol.Chem., 289, 2014
|
|
3WMB
 
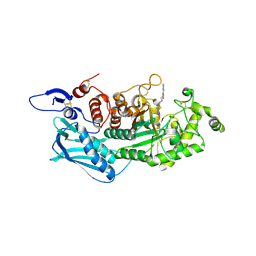 | | Crystal structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with naphthalimide derivative Q1 | | Descriptor: | 2-(2-{[(5-methyl-1,3,4-thiadiazol-2-yl)methyl]amino}ethyl)-1H-benzo[de]isoquinoline-1,3(2H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-hexosaminidase | | Authors: | Liu, T, Zhou, Y, Chen, L, Yang, Q. | | Deposit date: | 2013-11-16 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A crystal structure-guided rational design switching non-carbohydrate inhibitors' specificity between two beta-GlcNAcase homologs
Sci Rep, 4, 2014
|
|
2DWD
 
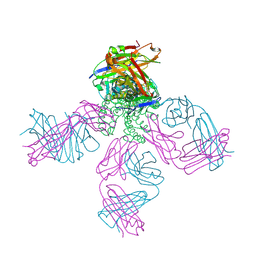 | | crystal structure of KcsA-FAB-TBA complex in Tl+ | | Descriptor: | (2S)-3-HYDROXY-2-(NONANOYLOXY)PROPYL LAURATE, ANTIBODY FAB HEAVY CHAIN, ANTIBODY FAB LIGHT CHAIN, ... | | Authors: | Yohannan, S, Zhou, Y. | | Deposit date: | 2006-08-10 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic Study of the Tetrabutylammonium Block to the KcsA K(+) Channel
J.Mol.Biol., 366, 2007
|
|
3VTR
 
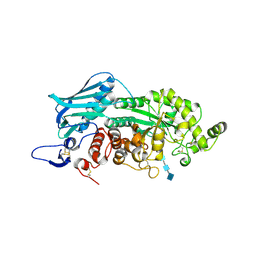 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 E328A complexed with TMG-chitotriomycin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-2-(trimethylammonio)-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetylglucosaminidase | | Authors: | Liu, T, Zhou, Y, Chen, L, Chen, W, Liu, L, Shen, X, Yang, Q. | | Deposit date: | 2012-06-02 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into cellulolytic and chitinolytic enzymes revealing crucial residues of insect beta-N-acetyl-D-hexosaminidase
Plos One, 7, 2012
|
|
1IJL
 
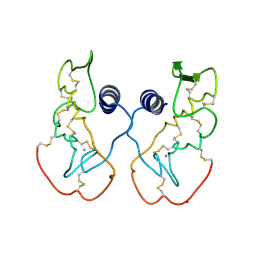 | | Crystal structure of acidic phospholipase A2 from deinagkistrodon acutus | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2, ZINC ION | | Authors: | Gu, L, Zhang, H, Song, S, Zhou, Y, Lin, Z. | | Deposit date: | 2001-04-27 | | Release date: | 2001-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of an acidic phospholipase A2 from the venom of Deinagkistrodon acutus.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3WZF
 
 | | Crystal structure of human cytoplasmic aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, cytoplasmic | | Authors: | Jiang, X, Chang, H, Zhou, Y, Chen, L, Yang, Q. | | Deposit date: | 2014-09-24 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.991 Å) | | Cite: | Recombinant expression, purification and Preliminary crystallographic studies of human cytoplasmic aspartate aminotransferase
To be Published
|
|
1M8S
 
 | | Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 5.9) | | Descriptor: | 1,4-BUTANEDIOL, CADMIUM ION, phospholipase a2 | | Authors: | Xu, S, Gu, L, Zhou, Y, Lin, Z. | | Deposit date: | 2002-07-25 | | Release date: | 2003-02-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of cadmium-binding acidic phospholipase A(2) from the venom of Agkistrodon halys Pallas at 1.9A resolutio
Biochem.Biophys.Res.Commun., 300, 2003
|
|
1M8R
 
 | | Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 7.4) | | Descriptor: | 1,4-BUTANEDIOL, CADMIUM ION, phospholipase A2 | | Authors: | Xu, S, Gu, L, Zhou, Y, Lin, Z. | | Deposit date: | 2002-07-25 | | Release date: | 2003-02-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of cadmium-binding acidic phospholipase A(2) from the venom of Agkistrodon halys Pallas at 1.9A resolutio
Biochem.Biophys.Res.Commun., 300, 2003
|
|
3W4R
 
 | | Crystal structure of an insect chitinase from the Asian corn borer, Ostrinia furnacalis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase | | Authors: | Chen, L, Liu, T, Zhou, Y, Shen, X, Yang, Q. | | Deposit date: | 2013-01-10 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characteristics of an insect group I chitinase, an enzyme indispensable to moulting
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WL0
 
 | | Crystal structure of Ostrinia furnacalis Group I chitinase catalytic domain E148A mutant in complex with a(GlcNAc)2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase | | Authors: | Chen, L, Liu, T, Zhou, Y, Chen, Q, Shen, X, Yang, Q. | | Deposit date: | 2013-11-05 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural characteristics of an insect group I chitinase, an enzyme indispensable to moulting.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
