5I8C
 
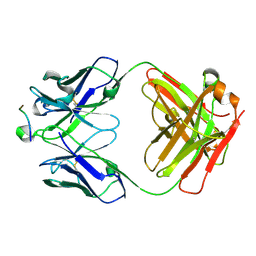 | | Crystal Structure of HIV-1 Clade A BG505 Fusion Peptide (residue 512-520) in Complex with Broadly Neutralizing Antibody VRC34.01 Fab | | Descriptor: | HIV-1 Clade A BG505 Fusion Peptide (residue 512-520), VRC34.01 Fab heavy chain, VRC34.01 Fab light chain | | Authors: | Xu, K, Zhou, T, Liu, K, Kwong, P.D. | | Deposit date: | 2016-02-18 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody.
Science, 352, 2016
|
|
3RJQ
 
 | | Crystal structure of anti-HIV llama VHH antibody A12 in complex with C186 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C186 gp120, Llama VHH A12 | | Authors: | Chen, L, McLellan, J.S, Kwon, Y.D, Schmidt, S, Wu, X, Zhou, T, Yang, Y, Zhang, B, Forsman, A, Weiss, R.A, Verrips, T, Mascola, J, Kwong, P.D. | | Deposit date: | 2011-04-15 | | Release date: | 2012-05-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Crystal structure of anti-HIV A12 VHH of llama antibody in complex with C1086 gp120
To be Published
|
|
5I8H
 
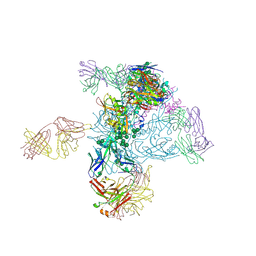 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer in Complex with V3 Loop-targeting Antibody PGT122 Fab and Fusion Peptide-targeting Antibody VRC34.01 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 SOSIP.664 gp120, ... | | Authors: | Xu, K, Zhou, T, Kwong, P.D. | | Deposit date: | 2016-02-18 | | Release date: | 2016-05-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.301 Å) | | Cite: | Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody.
Science, 352, 2016
|
|
3MME
 
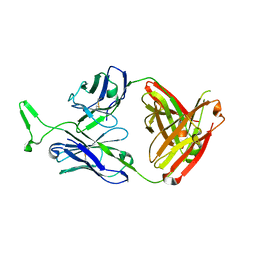 | | Structure and functional dissection of PG16, an antibody with broad and potent neutralization of HIV-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PG16 HEAVY CHAIN FAB, ... | | Authors: | Pancera, M, McLellan, J, Zhou, T, Zhu, J, Kwong, P. | | Deposit date: | 2010-04-19 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.97 Å) | | Cite: | Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1.
J.Virol., 84, 2010
|
|
3R6J
 
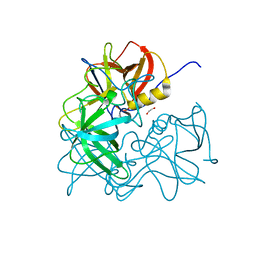 | | Crystal Structure of the Capsid P Domain from Norwalk Virus Strain Hiroshima/1999 | | Descriptor: | 1,2-ETHANEDIOL, VP1 protein | | Authors: | Hansman, G.S, Biertumpfel, C, McLellan, J.S, Georgiev, I, Chen, L, Zhou, T, Katayama, K, Kwong, P.D. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of GII.10 and GII.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability.
J.Virol., 85, 2011
|
|
4JB9
 
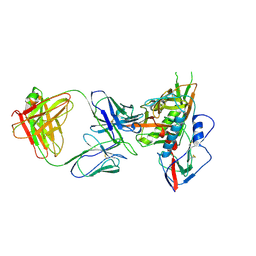 | | Crystal structure of antibody VRC06 in complex with HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, antibody VRC06 heavy chain, antibody VRC06 light chain, ... | | Authors: | Kwon, Y.D, Zhou, T, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2013-02-19 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization.
Science, 340, 2013
|
|
3R6K
 
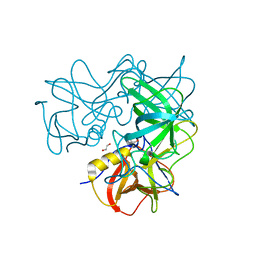 | | Crystal Structure of the Capsid P Domain from Norwalk Virus Strain Hiroshima/1999 in complex with HBGA type B (triglycan) | | Descriptor: | 1,2-ETHANEDIOL, VP1 protein, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Hansman, G.S, Biertumpfel, C, McLellan, J.S, Georgiev, I, Chen, L, Zhou, T, Katayama, K, Kwong, P.D. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of GII.10 and GII.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability.
J.Virol., 85, 2011
|
|
3SE9
 
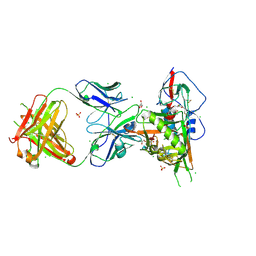 | | Crystal structure of broadly and potently neutralizing antibody VRC-PG04 in complex with HIV-1 gp120 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kwong, P.D, Zhou, T. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-10 | | Last modified: | 2021-04-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing.
Science, 333, 2011
|
|
3SE8
 
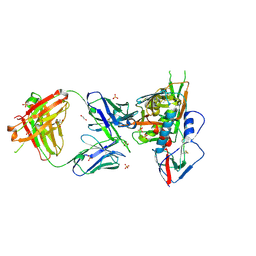 | | Crystal structure of broadly and potently neutralizing antibody VRC03 in complex with HIV-1 gp120 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kwong, P.D, Zhou, T. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-10 | | Last modified: | 2021-04-07 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing.
Science, 333, 2011
|
|
4YE4
 
 | | Crystal Structure of Neutralizing Antibody HJ16 in Complex with HIV-1 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HT593.1 gp120, Heavy chain human antibody HJ16, ... | | Authors: | Kwong, P.D, Chen, L, Zhou, T. | | Deposit date: | 2015-02-23 | | Release date: | 2015-07-22 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell, 161, 2015
|
|
4YFL
 
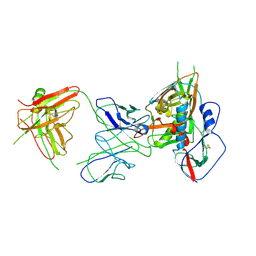 | | Crystal structure of VH1-46 germline-derived CD4-binding site-directed antibody 1B2530 in complex with HIV-1 clade A/E 93TH057 gp120 | | Descriptor: | 1B2530 Light chain, 1B2530 heavy chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Acharya, P, Zhou, T, Moquin, S, Kwong, P.D. | | Deposit date: | 2015-02-25 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.387 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell, 161, 2015
|
|
2QD0
 
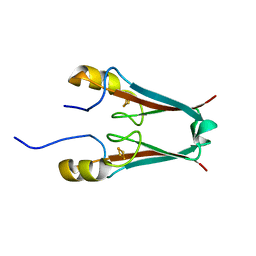 | | Crystal structure of mitoNEET | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Zinc finger CDGSH domain-containing protein 1 | | Authors: | Lin, J, Zhou, T, Ye, K, Wang, J. | | Deposit date: | 2007-06-20 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of human mitoNEET reveals distinct groups of iron sulfur proteins.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3HI1
 
 | | Structure of HIV-1 gp120 (core with V3) in Complex with CD4-Binding-Site Antibody F105 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F105 Heavy Chain, F105 Light Chain, ... | | Authors: | Kwon, Y.D, Chen, L, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z, Zhang, M.-Y, Arthos, J, Burton, D.R, Dimitrov, D, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-05-18 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
3IDX
 
 | | Crystal structure of HIV-gp120 core in complex with CD4-binding site antibody b13, space group C222 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fab b13 heavy chain, ... | | Authors: | Chen, L, Kwon, Y.D, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z.Y, Zhang, M.Y, Arthos, J, Burton, D.R, Dimitrov, D.S, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
3IDY
 
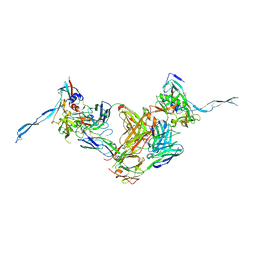 | | Crystal structure of HIV-gp120 core in complex with CD4-binding site antibody b13, space group C2221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab b13 heavy chain, Fab b13 light chain, ... | | Authors: | Chen, L, Kwon, Y.D, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z.Y, Zhang, M.Y, Arthos, J, Burton, D.R, Dimitrov, D.S, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
3JWO
 
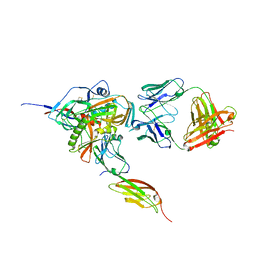 | | Structure of HIV-1 gp120 with gp41-Interactive Region: Layered Architecture and Basis of Conformational Mobility | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB 48D Heavy CHAIN, FAB 48D LIGHT CHAIN, ... | | Authors: | Pancera, M, Majeed, S, Huang, C.C, Kwon, Y.D, Zhou, T, Robinson, J.E, Sodroski, J, Wyatt, R, Kwong, P.D. | | Deposit date: | 2009-09-18 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3JWD
 
 | | Structure of HIV-1 gp120 with gp41-Interactive Region: Layered Architecture and Basis of Conformational Mobility | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB 48D HEAVY CHAIN, FAB 48D LIGHT CHAIN, ... | | Authors: | Pancera, M, Majeed, S, Ban, Y.A, Chen, L, Huang, C.C, Kong, L, Kwon, Y.D, Stuckey, J, Zhou, T, Robinson, J.E, Schief, W.R, Sodroski, J, Wyatt, R, Kwong, P.D. | | Deposit date: | 2009-09-18 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6USV
 
 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with glycine and SDZ 220-040 | | Descriptor: | (2S)-2-amino-3-[2',4'-dichloro-4-hydroxy-5-(phosphonomethyl)biphenyl-3-yl]propanoic acid, GLYCEROL, GLYCINE, ... | | Authors: | Romero-Hernandez, A, Tajima, N, Chou, T, Furukawa, h. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6USU
 
 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with L689,560 and glutamate | | Descriptor: | (2R,4S)-5,7-dichloro-4-[(phenylcarbamoyl)amino]-1,2,3,4-tetrahydroquinoline-2-carboxylic acid, GLUTAMIC ACID, Glutamate receptor ionotropic, ... | | Authors: | Romero-Hernandez, A, Tajima, N, Chou, T, Furukawa, H. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6ISL
 
 | | SnoaL-like cyclase XimE with its product xiamenmycin B | | Descriptor: | (2R,3S)-2-methyl-2-(4-methylpent-3-enyl)-3-oxidanyl-3,4-dihydrochromene-6-carboxylic acid, XimE, SnoaL-like domain protein | | Authors: | He, B, Zhou, T, Bu, X, Weng, J, Xu, J, Lin, S, Zheng, J, Zhao, Y, Xu, M. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Enzymatic Pyran Formation Involved in Xiamenmycin Biosynthesis
Acs Catalysis, 9, 2019
|
|
6ISK
 
 | | SnoaL-like cyclase XimE | | Descriptor: | 1,2-ETHANEDIOL, MALONIC ACID, XimE, ... | | Authors: | He, B, Zhou, T, Bu, X, Weng, J, Xu, J, Lin, S, Zheng, J, Zhao, Y, Xu, M. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Enzymatic Pyran Formation Involved in Xiamenmycin Biosynthesis
Acs Catalysis, 9, 2019
|
|
1H74
 
 | | CRYSTAL STRUCTURE OF HOMOSERINE KINASE COMPLEXED WITH ILE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HOMOSERINE KINASE, ISOLEUCINE, ... | | Authors: | Krishna, S.S, Zhou, T, Daugherty, M, Osterman, A.L, Zhang, H. | | Deposit date: | 2001-07-02 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Catalysis and Substrate Specificity of Homoserine Kinase
Biochemistry, 40, 2001
|
|
4D4E
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | Descriptor: | ARMADILLO REPEAT PROTEIN ARM00016, GLYCEROL | | Authors: | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | Deposit date: | 2014-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
4D49
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | Descriptor: | ARGININE, ARMADILLO REPEAT PROTEIN ARM00027, POLY ARG DECAPEPTIDE | | Authors: | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | Deposit date: | 2014-10-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
1PS5
 
 | | STRUCTURE OF THE MONOCLINIC C2 FORM OF HEN EGG-WHITE LYSOZYME AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | Lysozyme C, SULFATE ION | | Authors: | Majeed, S, Ofek, G, Belachew, A, Huang, C, Zhou, T, Kwong, P.D. | | Deposit date: | 2003-06-20 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enhancing Protein Crystallization through Precipitant Synergy
Structure, 11, 2003
|
|
