6INE
 
 | | Crystal Structure of human ASH1L-MRG15 complex | | Descriptor: | GLYCEROL, Histone-lysine N-methyltransferase ASH1L, Mortality factor 4-like protein 1, ... | | Authors: | Hou, P, Huang, C, Liu, C.P, Yu, T, Yin, Y, Zhu, B, Xu, R.M. | | Deposit date: | 2018-10-25 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into Stimulation of Ash1L's H3K36 Methyltransferase Activity through Mrg15 Binding.
Structure, 27, 2019
|
|
3VM7
 
 | | Structure of an Alpha-Amylase from Malbranchea cinnamomea | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Zhou, P, Hu, S.Q, Zhou, Y, Han, P, Yang, S.Q, Jiang, Z.Q. | | Deposit date: | 2011-12-09 | | Release date: | 2013-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Novel Multifunctional alpha-Amylase from the Thermophilic Fungus Malbranchea cinnamomea: Biochemical Characterization and Three-Dimensional Structure.
Appl Biochem Biotechnol., 170, 2013
|
|
8UKQ
 
 | | RNA polymerase II elongation complex with Fapy-dG lesion in apo state | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Hou, P, Oh, J, Wang, D. | | Deposit date: | 2023-10-15 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular Mechanism of RNA Polymerase II Transcriptional Mutagenesis by the Epimerizable DNA Lesion, Fapy·dG.
J.Am.Chem.Soc., 146, 2024
|
|
8UKT
 
 | | RNA polymerase II elongation complex with Fapy-dG lesion with AMP added | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Hou, P, Oh, J, Wang, D. | | Deposit date: | 2023-10-15 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Molecular Mechanism of RNA Polymerase II Transcriptional Mutagenesis by the Epimerizable DNA Lesion, Fapy·dG.
J.Am.Chem.Soc., 146, 2024
|
|
8UKU
 
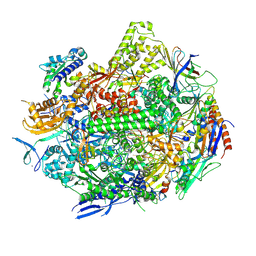 | | RNA polymerase II elongation complex with Fapy-dG lesion with CMP added | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Hou, P, Oh, J, Wang, D. | | Deposit date: | 2023-10-15 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Molecular Mechanism of RNA Polymerase II Transcriptional Mutagenesis by the Epimerizable DNA Lesion, Fapy·dG.
J.Am.Chem.Soc., 146, 2024
|
|
8UKS
 
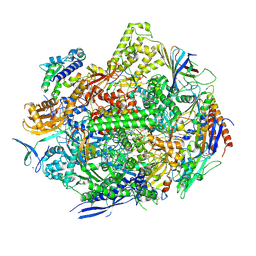 | | RNA polymerase II elongation complex with Fapy-dG lesion soaking with CTP before chemistry | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Hou, P, Oh, J, Wang, D. | | Deposit date: | 2023-10-15 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular Mechanism of RNA Polymerase II Transcriptional Mutagenesis by the Epimerizable DNA Lesion, Fapy·dG.
J.Am.Chem.Soc., 146, 2024
|
|
8UKR
 
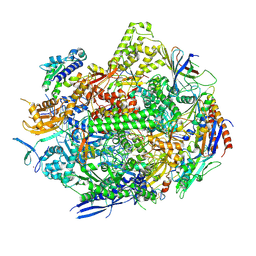 | | RNA polymerase II elongation complex with Fapy-dG lesion soaking with ATP before chemistry | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Hou, P, Oh, J, Wang, D. | | Deposit date: | 2023-10-15 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.78 Å) | | Cite: | Molecular Mechanism of RNA Polymerase II Transcriptional Mutagenesis by the Epimerizable DNA Lesion, Fapy·dG.
J.Am.Chem.Soc., 146, 2024
|
|
7FEX
 
 | |
7OX8
 
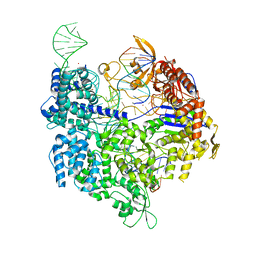 | | Target-bound SpCas9 complex with TRAC full RNA guide | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
7OXA
 
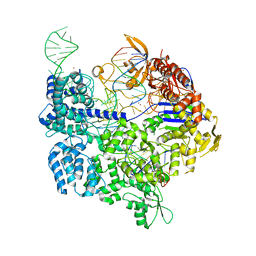 | | Target-bound SpCas9 complex with AAVS1 chimeric RNA-DNA guide | | Descriptor: | AAVS1 non-target DNA strand, AAVS1 target DNA strand, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
7OX7
 
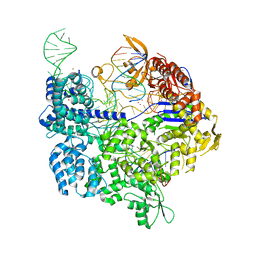 | | Target-bound SpCas9 complex with TRAC chimeric RNA-DNA guide | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
1IBX
 
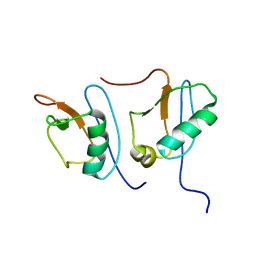 | | NMR STRUCTURE OF DFF40 AND DFF45 N-TERMINAL DOMAIN COMPLEX | | Descriptor: | CHIMERA OF IGG BINDING PROTEIN G AND DNA FRAGMENTATION FACTOR 45, DNA FRAGMENTATION FACTOR 40 | | Authors: | Zhou, P, Lugovskoy, A.A, McCarty, J.S, Li, P, Wagner, G. | | Deposit date: | 2001-03-29 | | Release date: | 2001-05-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of DFF40 and DFF45 N-terminal domain complex and mutual chaperone activity of DFF40 and DFF45.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2I5O
 
 | |
1C15
 
 | | SOLUTION STRUCTURE OF APAF-1 CARD | | Descriptor: | APOPTOTIC PROTEASE ACTIVATING FACTOR 1 | | Authors: | Zhou, P, Chou, J, Olea, R.S, Yuan, J, Wagner, G. | | Deposit date: | 1999-07-20 | | Release date: | 1999-09-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Apaf-1 CARD and its interaction with caspase-9 CARD: a structural basis for specific adaptor/caspase interaction.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1A66
 
 | | SOLUTION NMR STRUCTURE OF THE CORE NFATC1/DNA COMPLEX, 18 STRUCTURES | | Descriptor: | CORE NFATC1, DNA (5'-D(*CP*AP*AP*TP*TP*TP*TP*CP*CP*TP*CP*G)-3'), DNA (5'-D(*CP*GP*AP*GP*GP*AP*AP*AP*AP*TP*TP*G)-3') | | Authors: | Zhou, P, Sun, L.J, Doetsch, V, Wagner, G, Verdine, G.L. | | Deposit date: | 1998-03-06 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the core NFATC1/DNA complex.
Cell(Cambridge,Mass.), 92, 1998
|
|
7WYO
 
 | | Structure of the EV71 3Cpro with 338 inhibitor | | Descriptor: | 3C protein, N-methyl-N-(4,5,6,7-tetrahydro-1,3-benzothiazol-2-ylmethyl)prop-2-enamide | | Authors: | Qin, B, Hou, P, Gao, X, Cui, S. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Acrylamide fragment inhibitors that induce unprecedented conformational distortions in enterovirus 71 3C and SARS-CoV-2 main protease.
Acta Pharm Sin B, 12, 2022
|
|
7WYL
 
 | | Structure of the EV71 3Cpro with 337 inhibitor | | Descriptor: | 3C protein, N-methyl-N-[[4-(trifluoromethyl)-1,3-thiazol-2-yl]methyl]prop-2-enamide | | Authors: | Qin, B, Hou, P, Gao, X, Cui, S. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Acrylamide fragment inhibitors that induce unprecedented conformational distortions in enterovirus 71 3C and SARS-CoV-2 main protease.
Acta Pharm Sin B, 12, 2022
|
|
7WYM
 
 | | Structure of the SARS-COV-2 main protease with 337 inhibitor | | Descriptor: | 3C-like proteinase nsp5, N-methyl-N-[[4-(trifluoromethyl)-1,3-thiazol-2-yl]methyl]prop-2-enamide | | Authors: | Qin, B, Hou, P, Gao, X, Cui, S. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Acrylamide fragment inhibitors that induce unprecedented conformational distortions in enterovirus 71 3C and SARS-CoV-2 main protease.
Acta Pharm Sin B, 12, 2022
|
|
7WYP
 
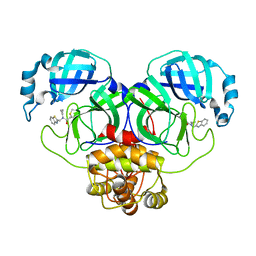 | | Structure of the SARS-COV-2 main protease with EN102 inhibitor | | Descriptor: | 3C-like proteinase, N-(1,3-benzothiazol-2-ylmethyl)-N-cyclopropyl-prop-2-enamide | | Authors: | Qin, B, Hou, P, Gao, X, Cui, S. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acrylamide fragment inhibitors that induce unprecedented conformational distortions in enterovirus 71 3C and SARS-CoV-2 main protease.
Acta Pharm Sin B, 12, 2022
|
|
6LAN
 
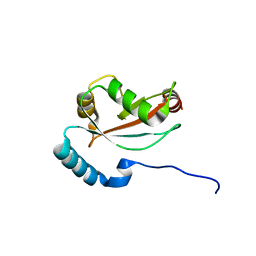 | | Structure of CCDC50 and LC3B complex | | Descriptor: | Coiled-coil domain-containing protein 50,Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Liu, L, Li, J, Hou, P. | | Deposit date: | 2019-11-12 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A novel selective autophagy receptor, CCDC50, delivers K63 polyubiquitination-activated RIG-I/MDA5 for degradation during viral infection.
Cell Res., 31, 2021
|
|
6NE8
 
 | |
9BD3
 
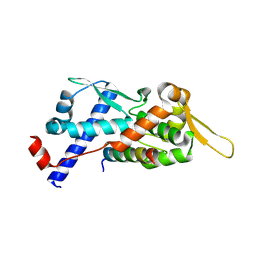 | | Structure of the MAGEA4 MHD-RAD18 R6BD Complex | | Descriptor: | E3 ubiquitin-protein ligase RAD18, Melanoma antigen A 4 | | Authors: | Forker, K, Zhou, P. | | Deposit date: | 2024-04-10 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of MAGEA4 MHD-RAD18 R6BD reveals a flipped binding mode compared to AlphaFold2 prediction.
Embo J., 2024
|
|
5DUR
 
 | | Influenza A virus H5 hemagglutinin globular head in complex with antibody 100F4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Antibody 100F4, Hemagglutinin, ... | | Authors: | Zuo, T, Sun, J, Wang, G, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2015-09-20 | | Release date: | 2015-12-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection
Nat Commun, 6, 2015
|
|
5YY5
 
 | | Structural definition of a unique neutralization epitope on the receptor-binding domain of MERS-CoV spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, Light chain, ... | | Authors: | Zhang, S, Wang, P, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2017-12-08 | | Release date: | 2018-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Definition of a Unique Neutralization Epitope on the Receptor-Binding Domain of MERS-CoV Spike Glycoprotein
Cell Rep, 24, 2018
|
|
7MK2
 
 | | CryoEM Structure of NPR1 | | Descriptor: | Regulatory protein NPR1, ZINC ION | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M, Bartesaghi, A, Zhou, P. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
