5X4H
 
 | | The crystal structure of Pyrococcus furiosus RecJ (wild-type) | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Li, M.J, Yi, G.S, Yu, F, Zhou, H, Chen, J.N, Xu, C.Y, Wang, F.P, Xiao, X, He, J.H, Liu, X.P. | | Deposit date: | 2017-02-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45.
Nucleic Acids Res., 45, 2017
|
|
5X4I
 
 | | Pyrococcus furiosus RecJ (D83A, Mn-soaking) | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Uncharacterized protein | | Authors: | Li, M.J, Yi, G.S, Yu, F, Zhou, H, Chen, J.N, Xu, C.Y, Wang, F.P, Xiao, X, He, J.H, Liu, X.P. | | Deposit date: | 2017-02-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45.
Nucleic Acids Res., 45, 2017
|
|
5X8Y
 
 | | A Mutation identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in vitro | | Descriptor: | ZIKV NS1 | | Authors: | Wang, D, Chen, C, Liu, S, Zhou, H, Yang, K, Zhao, Q, Ji, X, Chen, C, Xie, W, Wang, Z, Mi, L.Z, Yang, H. | | Deposit date: | 2017-03-03 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.817 Å) | | Cite: | A Mutation Identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in Vitro
Sci Rep, 7, 2017
|
|
7CBB
 
 | |
7CJY
 
 | |
6C29
 
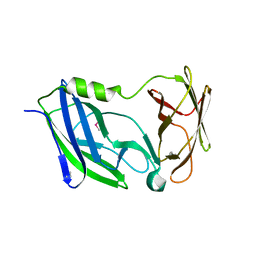 | | Crystal structure of the N-terminal periplasmic domain of ScsB from Proteus mirabilis | | Descriptor: | Putative metal resistance protein | | Authors: | Furlong, E.J, Choudhury, H.G, Kurth, F, Martin, J.L. | | Deposit date: | 2018-01-07 | | Release date: | 2018-03-07 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.538 Å) | | Cite: | Disulfide isomerase activity of the dynamic, trimericProteus mirabilisScsC protein is primed by the tandem immunoglobulin-fold domain of ScsB.
J. Biol. Chem., 293, 2018
|
|
2MMU
 
 | | Structure of CrgA, a Cell Division Structural and Regulatory Protein from Mycobacterium tuberculosis, in Lipid Bilayers | | Descriptor: | Cell division protein CrgA | | Authors: | Das, N, Dai, J, Hung, I, Rajagopalan, M, Zhou, H, Cross, T.A. | | Deposit date: | 2014-03-18 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure of CrgA, a cell division structural and regulatory protein from Mycobacterium tuberculosis, in lipid bilayers.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZN1
 
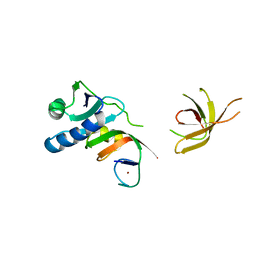 | | Crystal Structure of MjSpt4:Spt5 complex conformation A | | Descriptor: | Transcription elongation factor Spt4, Transcription elongation factor Spt5, ZINC ION | | Authors: | Guo, G.R, Zhou, H.H, Gao, Y.X, Zhu, Z.L, Niu, L.W, Teng, M.K. | | Deposit date: | 2015-05-04 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical insights into the DNA-binding mode of MjSpt4p:Spt5 complex at the exit tunnel of RNAPII
J.Struct.Biol., 192, 2015
|
|
5ZBA
 
 | | Crystal structure of Rtt109-Asf1-H3-H4-CoA complex | | Descriptor: | COENZYME A, DNA damage response protein Rtt109, putative, ... | | Authors: | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | Deposit date: | 2018-02-10 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
5Z9E
 
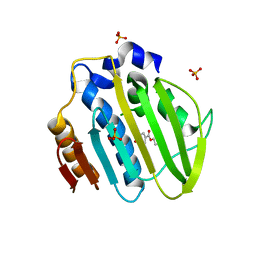 | |
5Z9M
 
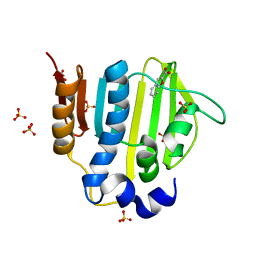 | |
5Z9B
 
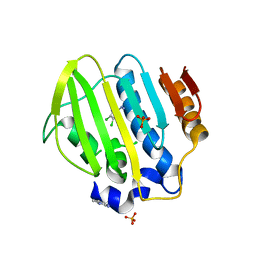 | | Bacterial GyrB ATPase domain in complex with (3,4-dichlorophenyl)hydrazine | | Descriptor: | (3,4-dichlorophenyl)hydrazine, 1H-benzimidazol-2-amine, DNA gyrase subunit B, ... | | Authors: | Huang, X, Zhou, H. | | Deposit date: | 2018-02-02 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of an auxiliary druggable pocket in the DNA gyrase ATPase domain using fragment probes
Medchemcomm, 9, 2018
|
|
5Z4H
 
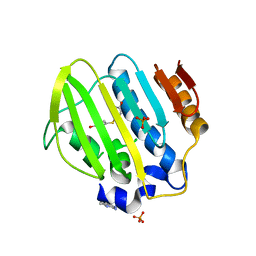 | | Bacterial GyrB ATPase domain in complex with a chemical fragment | | Descriptor: | 1H-benzimidazol-2-amine, 4-(4-hydroxyphenyl)sulfanylphenol, DNA gyrase subunit B, ... | | Authors: | Huang, X, Zhou, H. | | Deposit date: | 2018-01-11 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of an auxiliary druggable pocket in the DNA gyrase ATPase domain using fragment probes
Medchemcomm, 9, 2018
|
|
5ZYQ
 
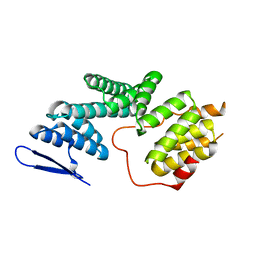 | | The Structure of Human PAF1/CTR9 complex | | Descriptor: | RNA polymerase-associated protein CTR9 homolog,RNA polymerase II-associated factor 1 homolog | | Authors: | Xie, Y, Zheng, M, Zhou, H, Long, J. | | Deposit date: | 2018-05-28 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.531 Å) | | Cite: | Paf1 and Ctr9 subcomplex formation is essential for Paf1 complex assembly and functional regulation.
Nat Commun, 9, 2018
|
|
5ZBB
 
 | | Crystal structure of Rtt109-Asf1-H3-H4 complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA damage response protein Rtt109, putative, ... | | Authors: | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | Deposit date: | 2018-02-10 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
5Z9P
 
 | |
5ZYP
 
 | | Structure of the Yeast Ctr9/Paf1 complex | | Descriptor: | NICKEL (II) ION, RNA polymerase-associated protein CTR9,RNA polymerase II-associated protein 1 | | Authors: | Xie, Y, Zheng, M, Zhou, H, Long, J. | | Deposit date: | 2018-05-28 | | Release date: | 2018-09-26 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (2.532 Å) | | Cite: | Paf1 and Ctr9 subcomplex formation is essential for Paf1 complex assembly and functional regulation.
Nat Commun, 9, 2018
|
|
7F43
 
 | |
7F42
 
 | |
5Z9F
 
 | | Bacterial GyrB ATPase domain in complex with a chemical fragment | | Descriptor: | 1H-benzimidazol-2-amine, 2-hydroxybenzonitrile, DNA gyrase subunit B, ... | | Authors: | Huang, X, Zhou, H. | | Deposit date: | 2018-02-03 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Identification of an auxiliary druggable pocket in the DNA gyrase ATPase domain using fragment probes
Medchemcomm, 9, 2018
|
|
5Z9L
 
 | | Bacterial GyrB ATPase domain in complex with a chemical fragment | | Descriptor: | 1H-benzimidazol-2-amine, 2-fluoro-4-hydroxybenzonitrile, DNA gyrase subunit B, ... | | Authors: | Huang, X, Zhou, H. | | Deposit date: | 2018-02-03 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of an auxiliary druggable pocket in the DNA gyrase ATPase domain using fragment probes
Medchemcomm, 9, 2018
|
|
5Z9Q
 
 | | Bacterial GyrB ATPase domain in complex with a chemical fragment | | Descriptor: | 1H-benzimidazol-2-amine, 5-phenyl-1H-pyrazol-3-amine, DNA gyrase subunit B, ... | | Authors: | Huang, X, Zhou, H. | | Deposit date: | 2018-02-04 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of an auxiliary druggable pocket in the DNA gyrase ATPase domain using fragment probes
Medchemcomm, 9, 2018
|
|
5Z4O
 
 | |
2L0J
 
 | | Solid State NMR structure of the M2 proton channel from Influenza A Virus in hydrated lipid bilayer | | Descriptor: | Matrix protein 2 | | Authors: | Sharma, M, Yi, M, Dong, H, Qin, H, Peterson, E, Busath, D.D, Zhou, H.X, Cross, T.A. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Insight into the mechanism of the influenza a proton channel from a structure in a lipid bilayer.
Science, 330, 2010
|
|
2MC7
 
 | | Structure of Salmonella MgtR | | Descriptor: | Regulatory peptide | | Authors: | Jean-Francois, F, Dai, J, Yu, L, Myrick, A, Rubin, E, Fajer, P, Song, L, Zhou, H, Cross, T. | | Deposit date: | 2013-08-15 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Binding of MgtR, a Salmonella Transmembrane Regulatory Peptide, to MgtC, a Mycobacterium tuberculosis Virulence Factor: A Structural Study.
J.Mol.Biol., 426, 2014
|
|
