5W36
 
 | |
6VG2
 
 | |
6VGG
 
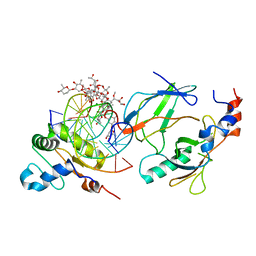 | | Crystal structure of the DNA binding domains of human transcription factor ERG, human Runx2 bound to core binding factor beta (Cbfb), and mithramycin, in complex with 16mer DNA CAGAGGATGTGGCTTC | | Descriptor: | Core-binding factor subunit beta, DNA (5'-D(P*CP*AP*GP*AP*GP*GP*AP*TP*GP*TP*GP*GP*CP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*GP*CP*CP*AP*CP*AP*TP*CP*CP*TP*CP*TP*G)-3'), ... | | Authors: | Hou, C, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2020-01-08 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.31 Å) | | Cite: | Allosteric interference in oncogenic FLI1 and ERG transactions by mithramycins.
Structure, 29, 2021
|
|
6VG8
 
 | |
5W34
 
 | |
5E8G
 
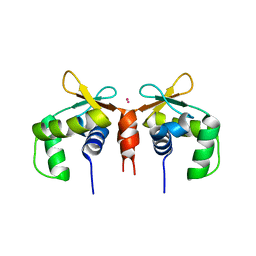 | |
5U9N
 
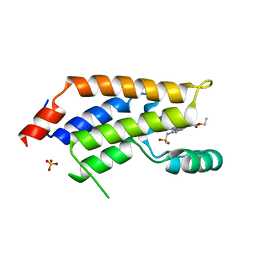 | | Second Bromodomain of cdg4_1340 from Cryptosporidium parvum, complexed with bromosporine | | Descriptor: | Bromo domain containing protein, Bromosporine, SULFATE ION | | Authors: | Hou, C.F.D, Lin, Y.H, Loppnau, P, Hutchinson, A, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, Walker, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-12-16 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Second Bromodomain of cdg4_1340 from Cryptosporidium parvum, complexed with bromosporine
To be published
|
|
5VS7
 
 | | Bromodomain of PF3D7_1475600 from Plasmodium falciparum complexed with peptide H4K5ac | | Descriptor: | Bromodomain protein, putative, CHLORIDE ION, ... | | Authors: | Hou, C.F.D, Loppnau, P, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, Walker, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-05-11 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Bromodomain of PF3D7_1475600 from Plasmodium falciparum complexed with peptide H4K5ac
To be published
|
|
5E8I
 
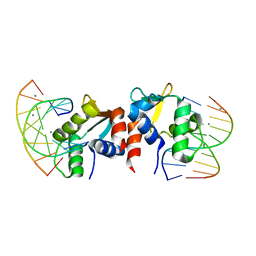 | |
4Z2Y
 
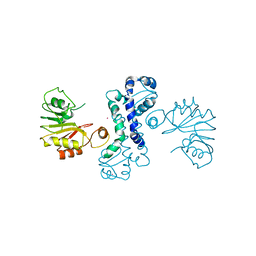 | | Crystal structure of methyltransferase CalO6 | | Descriptor: | CalO6, MERCURY (II) ION | | Authors: | Hou, C, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of O-methyltransferase CalO6 from the calicheamicin biosynthetic pathway: a case of challenging structure determination at low resolution.
Bmc Struct.Biol., 15, 2015
|
|
4P62
 
 | |
2JN5
 
 | |
7XKS
 
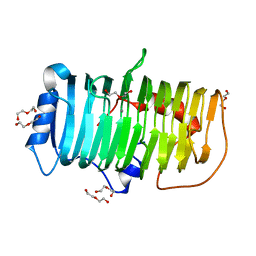 | | Crystal structure of an alkaline pectate lyase from Bacillus clausii | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Pectate lyase | | Authors: | Zhou, C, Zheng, Y.Y, Liu, W.D, Ma, Y. | | Deposit date: | 2022-04-20 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of an Alkaline Pectate Lyase and Rational Engineering with Improved Thermo-Alkaline Stability for Efficient Ramie Degumming.
Int J Mol Sci, 24, 2022
|
|
2KLZ
 
 | |
2JNH
 
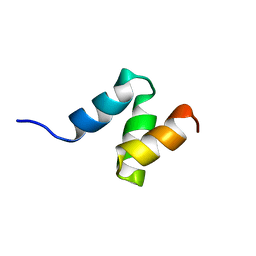 | | Solution Structure of the UBA Domain from Cbl-b | | Descriptor: | E3 ubiquitin-protein ligase CBL-B | | Authors: | Zhou, C, Zhou, Z, Lin, D, Hu, H. | | Deposit date: | 2007-01-24 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Differential ubiquitin binding of the UBA domains from human c-Cbl and Cbl-b: NMR structural and biochemical insights
Protein Sci., 17, 2008
|
|
7XT2
 
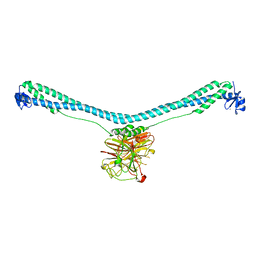 | | Crystal structure of TRIM72 | | Descriptor: | Tripartite motif-containing protein 72, ZINC ION | | Authors: | Zhou, C, Ma, Y.M, Ding, L. | | Deposit date: | 2022-05-15 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for TRIM72 oligomerization during membrane damage repair.
Nat Commun, 14, 2023
|
|
2LGW
 
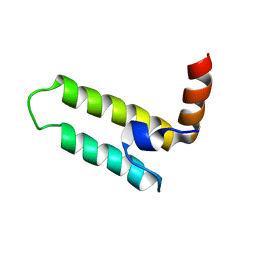 | | Solution Structure of the J Domain of HSJ1a | | Descriptor: | DnaJ homolog subfamily B member 2 | | Authors: | Zhou, C, Gao, X, Cao, C, Hu, H. | | Deposit date: | 2011-08-02 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C-terminal helices of heat shock protein 70 are essential for J-domain binding and ATPase activation.
J.Biol.Chem., 287, 2012
|
|
1XE8
 
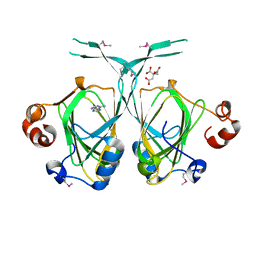 | | Crystal structure of the YML079w protein from Saccharomyces cerevisiae reveals a new sequence family of the jelly roll fold. | | Descriptor: | ADENINE, CITRIC ACID, GLYCEROL, ... | | Authors: | Zhou, C.-Z, Meyer, P, Quevillon-Cheruel, S, Li de La Sierra-Gallay, I, Collinet, B, Graille, M, Leulliot, N, Sorel, I, Janin, J, Van Tilbeurgh, H. | | Deposit date: | 2004-09-09 | | Release date: | 2005-01-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the YML079w protein from Saccharomyces cerevisiae reveals a new sequence family of the jelly-roll fold
Protein Sci., 14, 2005
|
|
2LMG
 
 | |
2JWQ
 
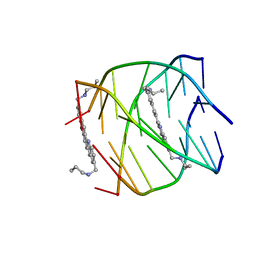 | | G-quadruplex recognition by quinacridines: a SAR, NMR and Biological study | | Descriptor: | DNA (5'-D(*DTP*DTP*DAP*DGP*DGP*DGP*DT)-3'), N,N'-(dibenzo[b,j][1,7]phenanthroline-2,10-diyldimethanediyl)dipropan-1-amine | | Authors: | Hounsou, C, Guittat, L, Monchaud, D, Jourdan, M, Saettel, N, Mergny, J.L, Teulade-Fichou, M. | | Deposit date: | 2007-10-23 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | G-Quadruplex Recognition by Quinacridines: a SAR, NMR, and Biological Study
ChemMedChem, 2, 2007
|
|
1XE7
 
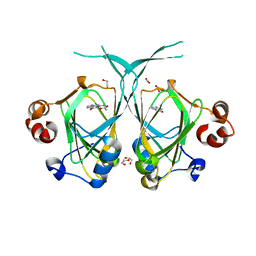 | | Crystal structure of the YML079w protein from Saccharomyces cerevisiae reveals a new sequence family of the jelly roll fold | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, GUANINE, ... | | Authors: | Zhou, C.-Z, Meyer, P, Quevillon-Cheruel, S, Li de La Sierra-Gallay, I, Collinet, B, Graille, M, Leulliot, N, Sorel, I, Janin, J, Van Tilbeurgh, H. | | Deposit date: | 2004-09-09 | | Release date: | 2005-01-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the YML079w protein from Saccharomyces cerevisiae reveals a new sequence family of the jelly-roll fold
Protein Sci., 14, 2005
|
|
8H8Y
 
 | | Crystal structure of AbHheG from Acidimicrobiia bacterium | | Descriptor: | GLYCEROL, alpha/beta hydrolase | | Authors: | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | Deposit date: | 2022-10-24 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
8HQP
 
 | | Crystal structure of AbHheG mutant from Acidimicrobiia bacterium | | Descriptor: | AbHheG_m | | Authors: | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | Deposit date: | 2022-12-13 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
5EAY
 
 | |
5EAN
 
 | | Crystal structure of Dna2 in complex with a 5' overhang DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, DNA (5'-D(P*AP*CP*TP*CP*TP*GP*CP*CP*AP*AP*GP*AP*GP*GP*A)-3'), ... | | Authors: | Zhou, C, Pourmal, S, Pavletich, N.P. | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Dna2 nuclease-helicase structure, mechanism and regulation by Rpa.
Elife, 4, 2015
|
|
