5I9J
 
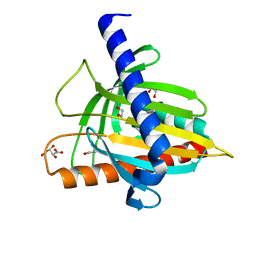 | | Structure of the cholesterol and lutein-binding domain of human STARD3 at 1.74A | | Descriptor: | 1,2-ETHANEDIOL, L(+)-TARTARIC ACID, SULFATE ION, ... | | Authors: | Horvath, M.P, Bernstein, P.S, Li, B, George, E.W, Tran, Q.T. | | Deposit date: | 2016-02-20 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure of the lutein-binding domain of human StARD3 at 1.74 angstrom resolution and model of a complex with lutein.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
1JB7
 
 | |
2I0Q
 
 | |
1OTC
 
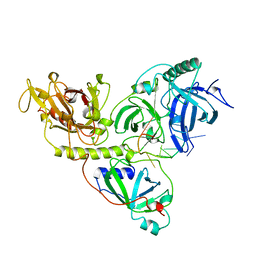 | | THE O. NOVA TELOMERE END BINDING PROTEIN COMPLEXED WITH SINGLE STRAND DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3'), PROTEIN (TELOMERE-BINDING PROTEIN ALPHA SUBUNIT), PROTEIN (TELOMERE-BINDING PROTEIN BETA SUBUNIT) | | Authors: | Horvath, M.P, Schweiker, V.L, Bevilacqua, J.M, Ruggles, J.A, Schultz, S.C. | | Deposit date: | 1998-11-25 | | Release date: | 1999-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA.
Cell(Cambridge,Mass.), 95, 1998
|
|
6U7T
 
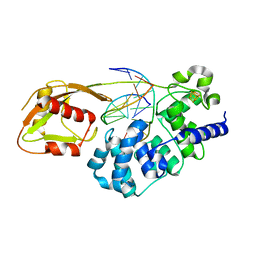 | | MutY adenine glycosylase bound to DNA containing a transition state analog (1N) paired with d(8-oxo-G) | | Descriptor: | Adenine DNA glycosylase, CALCIUM ION, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), ... | | Authors: | O'Shea Murray, V.L, Cao, S, Horvath, M.P, David, S.S. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Finding OG Lesions and Avoiding Undamaged G by the DNA Glycosylase MutY.
Acs Chem.Biol., 15, 2020
|
|
9BS2
 
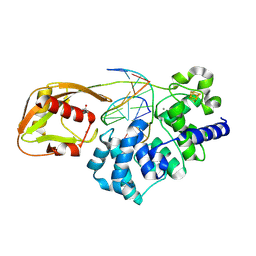 | | Glycosylase MutY variant R149Q in complex with DNA containing d(8-oxo-G) paired with a product analog (THF) to 1.51 A resolution | | Descriptor: | ACETIC ACID, Adenine DNA glycosylase, CALCIUM ION, ... | | Authors: | Trasvina-Arenas, C.H, Tamayo, N, Lin, W.J, Demir, M, Fisher, A.J, David, S.S, Horvath, M.P. | | Deposit date: | 2024-05-12 | | Release date: | 2024-10-30 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure of human MUTYH and functional profiling of cancer-associated variants reveal an allosteric network between its [4Fe-4S] cluster cofactor and active site required for DNA repair.
Nat Commun, 16, 2025
|
|
6Q0C
 
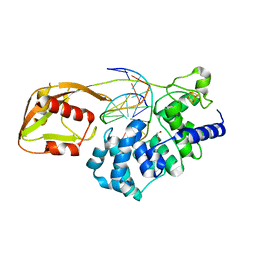 | | MutY adenine glycosylase bound to DNA containing a transition state analog (1N) paired with undamaged dG | | Descriptor: | 1,2-ETHANEDIOL, A/G-specific adenine glycosylase, CALCIUM ION, ... | | Authors: | O'Shea Murray, V.L, Russelburg, L.P, Horvath, M.P, David, S.S. | | Deposit date: | 2019-08-01 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Finding OG Lesions and Avoiding Undamaged G by the DNA Glycosylase MutY.
Acs Chem.Biol., 15, 2020
|
|
3FP6
 
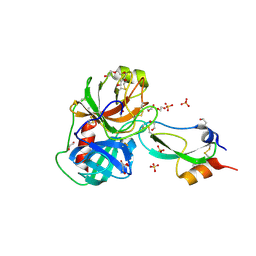 | | Anionic trypsin in complex with bovine pancreatic trypsin inhibitor (BPTI) determined to the 1.49 A resolution limit | | Descriptor: | 1,2-ETHANEDIOL, Anionic trypsin-2, CALCIUM ION, ... | | Authors: | Zakharova, E, Horvath, M.P, Goldenberg, D.P. | | Deposit date: | 2009-01-04 | | Release date: | 2009-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure of a serine protease poised to resynthesize a peptide bond.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3FP7
 
 | | Anionic trypsin variant S195A in complex with bovine pancreatic trypsin inhibitor (BPTI) cleaved at the scissile bond (LYS15-ALA16) determined to the 1.46 A resolution limit | | Descriptor: | 1,2-ETHANEDIOL, Anionic trypsin-2, CALCIUM ION, ... | | Authors: | Zakharova, E, Horvath, M.P, Goldenberg, D.P. | | Deposit date: | 2009-01-04 | | Release date: | 2009-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of a serine protease poised to resynthesize a peptide bond.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3FP8
 
 | | Anionic trypsin variant S195A in complex with bovine pancreatic trypsin inhibitor (BPTI) determined to the 1.46 A resolution limit | | Descriptor: | 1,2-ETHANEDIOL, Anionic trypsin-2, CALCIUM ION, ... | | Authors: | Zakharova, E, Horvath, M.P, Goldenberg, D.P, Curtice, K. | | Deposit date: | 2009-01-04 | | Release date: | 2009-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of a serine protease poised to resynthesize a peptide bond.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8FAY
 
 | | Human MUTYH adenine glycosylase bound to DNA containing a transition state analog (1N) paired with d(8-oxo-G) | | Descriptor: | Adenine DNA glycosylase, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), DNA (5'-D(P*GP*TP*CP*CP*AP*(NR1)P*GP*TP*CP*T)-3'), ... | | Authors: | Trasvina-Arenas, C.H, Lin, W.J, Demir, M, Fisher, A.J, David, S.S, Horvath, M.P. | | Deposit date: | 2022-11-29 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Human MUTYH adenine glycosylase bound to DNA containing a transition state analog (1N) paired with d(8-oxo-G)
To Be Published
|
|
8DVP
 
 | | Glycosylase MutY variant N146S in complex with DNA containing d(8-oxo-G) paired with substrate purine | | Descriptor: | ACETATE ION, Adenine DNA glycosylase, CALCIUM ION, ... | | Authors: | Demir, M, Russelburg, L.P, Horvath, M.P, David, S.S. | | Deposit date: | 2022-07-29 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural snapshots of base excision by the cancer-associated variant MutY N146S reveal a retaining mechanism.
Nucleic Acids Res., 51, 2023
|
|
8DW0
 
 | | Glycosylase MutY variant N146S in complex with DNA containing d(8-oxo-G) paired with an enzyme-generated abasic site (AP) product and crystallized with sodium acetate | | Descriptor: | 1,2-ETHANEDIOL, Adenine DNA glycosylase, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), ... | | Authors: | Demir, M, Russelburg, L.P, Horvath, M.P, David, S.S. | | Deposit date: | 2022-07-30 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural snapshots of base excision by the cancer-associated variant MutY N146S reveal a retaining mechanism.
Nucleic Acids Res., 51, 2023
|
|
8DW7
 
 | | DNA glycosylase MutY variant N146S in complex with DNA containing the transition state analog 1N paired with d(8-oxo-G) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adenine DNA glycosylase, ... | | Authors: | Demir, M, Russelburg, L.P, Horvath, M.P, David, S.S. | | Deposit date: | 2022-07-31 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural snapshots of base excision by the cancer-associated variant MutY N146S reveal a retaining mechanism.
Nucleic Acids Res., 51, 2023
|
|
8DVY
 
 | | DNA glycosylase MutY variant N146S in complex with DNA containing d(8-oxo-G) paired with an enzyme-generated abasic site product (AP) and crystalized with calcium acetate | | Descriptor: | ACETATE ION, Adenine DNA glycosylase, CALCIUM ION, ... | | Authors: | Demir, M, Russelburg, L.P, Horvath, M.P, David, S.S. | | Deposit date: | 2022-07-30 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural snapshots of base excision by the cancer-associated variant MutY N146S reveal a retaining mechanism.
Nucleic Acids Res., 51, 2023
|
|
8DW4
 
 | | Glycosylase MutY variant N146S in complex with DNA containing d(8-oxo-G) paired with an abasic site product (AP) generated by the enzyme in crystals by removal of calcium | | Descriptor: | ACETATE ION, Adenine DNA glycosylase, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), ... | | Authors: | Demir, M, Russelburg, L.P, Horvath, M.P, David, S.S. | | Deposit date: | 2022-07-31 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural snapshots of base excision by the cancer-associated variant MutY N146S reveal a retaining mechanism.
Nucleic Acids Res., 51, 2023
|
|
8DWD
 
 | | Adenine glycosylase MutY variant E43S in complex with DNA containing d(8-oxo-G) paired with an AP site generated by the enzyme acting on purine | | Descriptor: | ACETATE ION, Adenine DNA glycosylase, CALCIUM ION, ... | | Authors: | Russelburg, L.P, Demir, M, David, S.S, Horvath, M.P. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Basis for Base Engagement and Stereochemistry Revealed by Alteration of Catalytic Residue Glu43 in DNA Repair Glycosylase MutY
To Be Published
|
|
8DWF
 
 | | Glycosylase MutY variant E43S in complex with DNA containing d(8-oxo-G) paired with substrate adenine | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adenine DNA glycosylase, ... | | Authors: | Russelburg, L.P, Demir, M, David, S.S, Horvath, M.P. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Base Engagement and Stereochemistry Revealed by Alteration of Catalytic Residue Glu43 in DNA Repair Glycosylase MutY
To Be Published
|
|
8DWE
 
 | | Adenine glycosylase MutY variant E43Q in complex with DNA containing d(8-oxo-G) paired with substrate purine | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adenine DNA glycosylase, ... | | Authors: | Russelburg, L.P, Demir, M, David, S.S, Horvath, M.P. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Base Engagement and Stereochemistry Revealed by Alteration of Catalytic Residue Glu43 in DNA Repair Glycosylase MutY
To Be Published
|
|
1Y62
 
 | | A 2.4 crystal structure of conkunitzin-S1, a novel Kunitz-fold cone snail neurotoxin. | | Descriptor: | Conkunitzin-S1, SULFATE ION | | Authors: | Dy, C.Y, Buczek, P, Horvath, M.P. | | Deposit date: | 2004-12-03 | | Release date: | 2005-07-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of conkunitzin-S1, a neurotoxin and Kunitz-fold disulfide variant from cone snail.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2FI5
 
 | | Crystal structure of a BPTI variant (Cys38->Ser) in complex with trypsin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | Authors: | Zakharova, E, Horvath, M.P, Goldenberg, D.P. | | Deposit date: | 2005-12-27 | | Release date: | 2006-01-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Functional and structural roles of the Cys14-Cys38 disulfide of bovine pancreatic trypsin inhibitor.
J.Mol.Biol., 382, 2008
|
|
2FI3
 
 | | Crystal structure of a BPTI variant (Cys14->Ser, Cys38->Ser) in complex with trypsin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | Authors: | Zakharova, E, Horvath, M.P, Goldenberg, D.P. | | Deposit date: | 2005-12-27 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Functional and structural roles of the Cys14-Cys38 disulfide of bovine pancreatic trypsin inhibitor.
J.Mol.Biol., 382, 2008
|
|
2FTL
 
 | | Crystal structure of trypsin complexed with BPTI at 100K | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | Authors: | Hanson, W.M, Horvath, M.P, Goldenberg, D.P. | | Deposit date: | 2006-01-24 | | Release date: | 2006-02-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Rigidification of a Flexible Protease Inhibitor Variant upon Binding to Trypsin.
J.Mol.Biol., 366, 2007
|
|
2FI4
 
 | | Crystal structure of a BPTI variant (Cys14->Ser) in complex with trypsin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | Authors: | Zakharova, E, Horvath, M.P, Goldenberg, D.P. | | Deposit date: | 2005-12-27 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Functional and structural roles of the Cys14-Cys38 disulfide of bovine pancreatic trypsin inhibitor.
J.Mol.Biol., 382, 2008
|
|
2FTM
 
 | | Crystal structure of trypsin complexed with the BPTI variant (Tyr35->Gly) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | Authors: | Hanson, W.M, Horvath, M.P, Goldenberg, D.P. | | Deposit date: | 2006-01-24 | | Release date: | 2006-02-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Rigidification of a Flexible Protease Inhibitor Variant upon Binding to Trypsin.
J.Mol.Biol., 366, 2007
|
|
