8GBU
 
 | | Hepatitis B capsid Y132A mutant with compound AB-506 | | Descriptor: | (1-methyl-1H-1,2,4-triazol-3-yl)methyl {(1S)-4-[(3-chloro-4-fluorophenyl)carbamoyl]-7-fluoro-2,3-dihydro-1H-inden-1-yl}carbamate, 1,2-ETHANEDIOL, Capsid protein | | Authors: | Horanyi, P.S, Mayclin, S.J. | | Deposit date: | 2023-02-28 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis, and structure-activity relationship of a bicyclic HBV capsid assembly modulator chemotype leading to the identification of clinical candidate AB-506.
Bioorg.Med.Chem.Lett., 94, 2023
|
|
4PFZ
 
 | |
8DOF
 
 | | Pseudomonas aeruginosa MurC with WYH9-2-P - OSA_001044 | | Descriptor: | (2R)-2-cyclohexyl-2-[(4-{[5-(propan-2-yl)-1H-pyrazol-3-yl]amino}-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]ethan-1-ol, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Horanyi, P.S, Wang, Y, Todd, M.H, Abendroth, J, Edwards, T, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-07-13 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pseudomonas aeruginosa MurC with WYH9-2-P - OSA_001044
To Be Published
|
|
3RGM
 
 | | Crystal structure of spin-labeled BtuB T156R1 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MAGNESIUM ION, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, ... | | Authors: | Horanyi, P.S, Freed, D.M, Wiener, M.C, Cafiso, D.S. | | Deposit date: | 2011-04-08 | | Release date: | 2011-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Origin of Electron Paramagnetic Resonance Line Shapes on β-Barrel Membrane Proteins: The Local Solvation Environment Modulates Spin-Label Configuration
Biochemistry, 50, 2011
|
|
4TMD
 
 | | X-ray structure of Putative uncharacterized protein (Rv0999 ortholog) from Mycobacterium smegmatis | | Descriptor: | IODIDE ION, Uncharacterized protein | | Authors: | Horanyi, P.S, Dranow, D.M, Abendroth, J, Lorimer, D, Edwards, T, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-06-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of Putative uncharacterized protein (Rv0999 ortholog) from Mycobacterium smegmatis
To Be Published
|
|
4TWR
 
 | | Structure of UDP-glucose 4-epimerase from Brucella abortus | | Descriptor: | NAD binding site:NAD-dependent epimerase/dehydratase:UDP-glucose 4-epimerase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Horanyi, P.S, Abendroth, J, Lorimer, D, Edwards, T, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-07-01 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of UDP-glucose 4-epimerase from Brucella melitensis
To Be Published
|
|
6X9N
 
 | | Pseudomonas aeruginosa MurC with AZ5595 | | Descriptor: | (2R)-2-({4-[(5-tert-butyl-1-methyl-1H-pyrazol-3-yl)amino]-1H-pyrazolo[3,4-d]pyrimidin-6-yl}amino)-2-phenylethan-1-ol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Horanyi, P.S, Mayclin, S.J, Durand-Reville, T.F, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-06-03 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Pseudomonas aeruginosa MurC with AZ5595
To Be Published
|
|
6X9F
 
 | | Pseudomonas aeruginosa MurC with AZ8074 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Horanyi, P.S, Mayclin, S.J, Durand-Reville, T.F, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-06-02 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Pseudomonas aeruginosa MurC with AZ8074
To Be Published
|
|
5UUI
 
 | | Crystal Structure of Spin-Labeled T77C TNFa | | Descriptor: | S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, Tumor necrosis factor | | Authors: | Horanyi, P.S, Dranow, D.M, Ceska, T. | | Deposit date: | 2017-02-16 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Natural Conformational Sampling of Human TNF alpha Visualized by Double Electron-Electron Resonance.
Biophys. J., 113, 2017
|
|
6CKG
 
 | | D-glycerate 3-kinase from Cryptococcus neoformans var. grubii serotype A (H99 / ATCC 208821 / CBS 10515 / FGSC 9487) | | Descriptor: | 1,2-ETHANEDIOL, D-glycerate 3-kinase | | Authors: | Horanyi, P.S, Abendroth, J, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-02-28 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D-glycerate 3-kinase from Cryptococcus neoformans var. grubii serotype A (H99 / ATCC 208821 / CBS 10515 / FGSC 9487)
To be Published
|
|
6CAU
 
 | | UDP-N-acetylmuramate--alanine ligase from Acinetobacter baumannii AB5075-UW with AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, UDP-N-acetylmuramate--L-alanine ligase | | Authors: | Horanyi, P.S, Abendroth, J, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-01-31 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | UDP-N-acetylmuramate--alanine ligase from Acinetobacter baumannii AB5075-UW with AMPPNP
To be Published
|
|
6U94
 
 | | Structure of RND efflux system, outer membrane lipoprotein, NodT family from Burkholderia mallei ATCC 23344 | | Descriptor: | GLYCEROL, RND efflux system, outer membrane lipoprotein, ... | | Authors: | Horanyi, P.S, Fox III, D, Abendroth, J, Lorimer, D, Edwards, T, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-09-06 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of RND efflux system, outer membrane lipoprotein, NodT family from Burkholderia mallei ATCC 23344
To Be Published
|
|
3M8D
 
 | | Crystal structure of spin-labeled BtuB V10R1 with bound calcium and cyanocobalamin | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CALCIUM ION, CYANOCOBALAMIN, ... | | Authors: | Freed, D.M, Horanyi, P.S, Wiener, M.C, Cafiso, D.S. | | Deposit date: | 2010-03-17 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Conformational exchange in a membrane transport protein is altered in protein crystals.
Biophys.J., 99, 2010
|
|
3M8B
 
 | | Crystal structure of spin-labeled BtuB V10R1 in the apo state | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MAGNESIUM ION, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, ... | | Authors: | Freed, D.M, Horanyi, P.S, Wiener, M.C, Cafiso, D.S. | | Deposit date: | 2010-03-17 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Conformational exchange in a membrane transport protein is altered in protein crystals.
Biophys.J., 99, 2010
|
|
8UOA
 
 | | Structure of the synaptic vesicle protein 2A Luminal domain in complex with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody, Synaptic vesicle glycoprotein 2A, ... | | Authors: | Mittal, A, Martin, M.F, Levin, E, Adams, C, Yang, M, Ledecq, M, Horanyi, P.S, Coleman, J.A. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of synaptic vesicle protein 2A and 2B bound to anticonvulsants.
Nat.Struct.Mol.Biol., 2024
|
|
8UO9
 
 | | Structure of synaptic vesicle protein 2A in complex with a nanobody | | Descriptor: | (4S)-1-{[(4S)-2-(methoxymethyl)-6-(trifluoromethyl)imidazo[2,1-b][1,3,4]thiadiazol-5-yl]methyl}-4-(4,4,4-trifluorobutyl)pyrrolidin-2-one, 1,2-DIDECANOYL-SN-GLYCERO-3-[PHOSPHO-L-SERINE], 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mittal, A, Martin, M.F, Levin, E, Adams, C, Yang, M, Ledecq, M, Horanyi, P.S, Coleman, J.A. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of synaptic vesicle protein 2A and 2B bound to anticonvulsants.
Nat.Struct.Mol.Biol., 2024
|
|
8UO8
 
 | | Structure of synaptic vesicle protein 2B with padsevonil | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, (4R)-4-(2-chloro-2,2-difluoroethyl)-1-{[(4R)-2-(methoxymethyl)-6-(trifluoromethyl)imidazo[2,1-b][1,3,4]thiadiazol-5-yl]methyl}pyrrolidin-2-one, 1,2-DIDECANOYL-SN-GLYCERO-3-[PHOSPHO-L-SERINE], ... | | Authors: | Martin, M.F, Mittal, A, Levin, E, Adams, C, Yang, M, Ledecq, M, Horanyi, P.S, Coleman, J.A. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of synaptic vesicle protein 2A and 2B bound to anticonvulsants.
Nat.Struct.Mol.Biol., 2024
|
|
3RGN
 
 | | Crystal structure of spin-labeled BtuB W371R1 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MAGNESIUM ION, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, ... | | Authors: | Freed, D.M, Horanyi, P.S, Wiener, M.C, Cafiso, D.S. | | Deposit date: | 2011-04-08 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Origin of Electron Paramagnetic Resonance Line Shapes on β-Barrel Membrane Proteins: The Local Solvation Environment Modulates Spin-Label Configuration
Biochemistry, 50, 2011
|
|
1ZD0
 
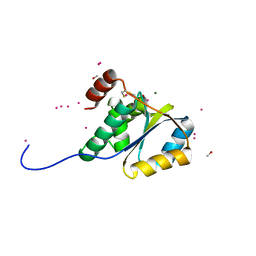 | | Crystal structure of Pfu-542154 conserved hypothetical protein | | Descriptor: | MAGNESIUM ION, METHANOL, UNKNOWN ATOM OR ION, ... | | Authors: | Habel, J.E, Liu, Z.J, Horanyi, P.S, Florence, Q.J.T, Tempel, W, Zhou, W, Chen, L, Lee, D, Nguyen, J, Chang, S.H, Bereton, P, Izumi, M, Jenny Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-04-13 | | Release date: | 2005-05-17 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Pfu-542154 conserved hypothetical protein
To be Published
|
|
4IL3
 
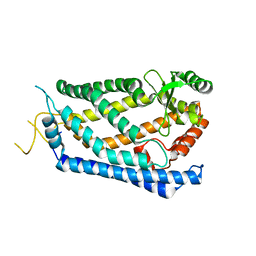 | | Crystal Structure of S. mikatae Ste24p | | Descriptor: | Ste24p, ZINC ION | | Authors: | Pryor Jr, E.E, Horanyi, P.S, Clark, K, Fedoriw, N, Connelly, S.M, Koszelak-Rosenblum, M, Zhu, G, Malkowski, M.G, Dumont, M.E, Wiener, M.C, Membrane Protein Structural Biology Consortium (MPSBC) | | Deposit date: | 2012-12-28 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structure of the integral membrane protein CAAX protease Ste24p.
Science, 339, 2013
|
|
4IRG
 
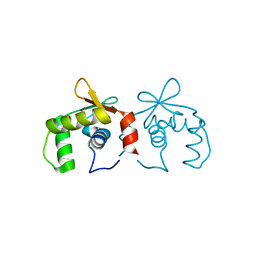 | | Uninhibited DNA-binding domain of the Ets transcription factor ERG | | Descriptor: | Transcriptional regulator ERG | | Authors: | Regan, M.C, Horanyi, P.S, Pryor, E.E, Sarver, J.L, Cafiso, D.S, Bushweller, J.H. | | Deposit date: | 2013-01-14 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and dynamic studies of the transcription factor ERG reveal DNA binding is allosterically autoinhibited.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4IRH
 
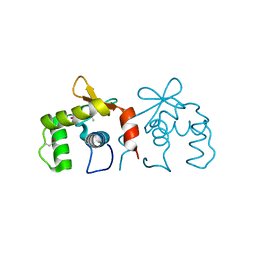 | | Auto-inhibited ERG Ets domain | | Descriptor: | Transcriptional regulator ERG | | Authors: | Regan, M.C, Horanyi, P.S, Pryor, E.E, Sarver, J.L, Cafiso, D.S, Bushweller, J.H. | | Deposit date: | 2013-01-14 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and dynamic studies of the transcription factor ERG reveal DNA binding is allosterically autoinhibited.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4IRI
 
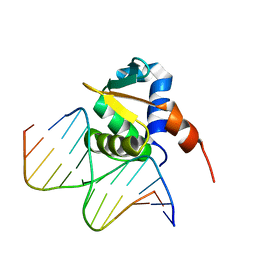 | | Auto-inhibited ERG Ets Domain-DNA Complex | | Descriptor: | DNA (5'-D(*CP*CP*AP*CP*TP*TP*CP*CP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP*G)-3'), Transcriptional regulator ERG | | Authors: | Regan, M.C, Horanyi, P.S, Pryor, E.E, Sarver, J.L, Cafiso, D.S, Bushweller, J.H. | | Deposit date: | 2013-01-14 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural and dynamic studies of the transcription factor ERG reveal DNA binding is allosterically autoinhibited.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7JWL
 
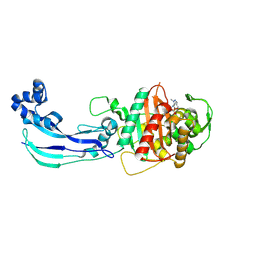 | | Crystal Structure of Pseudomonas aeruginosa Penicillin Binding Protein 3 (PAE-PBP3) bound to ETX0462 | | Descriptor: | CHLORIDE ION, ETX0462 (Bound form), Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Mayclin, S.J, Abendroth, J, Horanyi, P.S, Sylvester, M, Wu, X, Shapiro, A, Moussa, S, Durand-Reville, T.F. | | Deposit date: | 2020-08-25 | | Release date: | 2021-05-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational design of a new antibiotic class for drug-resistant infections.
Nature, 597, 2021
|
|
1UPS
 
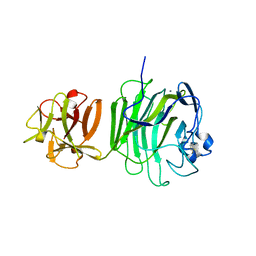 | | GlcNAc[alpha]1-4Gal releasing endo-[beta]-galactosidase from Clostridium perfringens | | Descriptor: | CALCIUM ION, GLCNAC-ALPHA-1,4-GAL-RELEASING ENDO-BETA-GALACTOSIDASE | | Authors: | Tempel, W, Liu, Z.-J, Horanyi, P.S, Deng, L, Lee, D, Newton, M.G, Rose, J.P, Ashida, H, Li, S.-C, Li, Y.-T, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-10-10 | | Release date: | 2004-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Three-dimensional structure of GlcNAcalpha1-4Gal releasing endo-beta-galactosidase from Clostridium perfringens.
Proteins, 59, 2005
|
|
